Hongyu Li1, Zifei Liang2, Chaoyi Zhang1, Ruiying Liu1, Jing Li3, Weihong Zhang3, Dong Liang4, Bowen Shen5, Peizhou Huang6, Sunil Kumar Gaire1, Xiaoliang Zhang6, Yulin Ge2, Jiangyang Zhang2, and Leslie Ying1,6
1Electrical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States, 2Center for Biomedical Imaging, Radiology, New York University School of Medicine, New York, NY, United States, 3Radiology, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China, 4Paul C. Lauterbur Research Center for Biomedical Imaging, Medical AI research center, SIAT, CAS, Shenzhen, China, 5Computer Science, Virginia Tech, Blacksburg, VA, United States, 6Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States
1Electrical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States, 2Center for Biomedical Imaging, Radiology, New York University School of Medicine, New York, NY, United States, 3Radiology, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China, 4Paul C. Lauterbur Research Center for Biomedical Imaging, Medical AI research center, SIAT, CAS, Shenzhen, China, 5Computer Science, Virginia Tech, Blacksburg, VA, United States, 6Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States
This paper demonstrates
the feasibility of superfast DTI and fiber tractography
using deep learning with as few as six corrupted
DWIs (up to 30-fold). Such a significant reduction in scan time will allow
the inclusion of DTI into clinical routine for many potential applications.
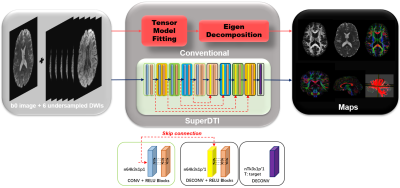
FIGURE
1. Schematic comparison of the conventional DTI model fitting and deep learning
methods SuperDTI for generating various diffusion quantification maps.
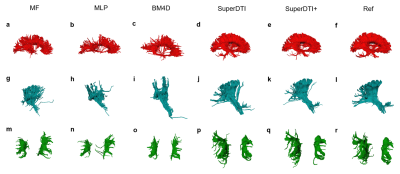
FIGURE 4. Comparison of fiber tractography generated from 6
DWIs using different methods. Corpus callosum, internal
capsule/corticospinal tract, and superior longitudinal fasciculus generated by
MF (a), MLP (b), BM4D (c), proposed SuperDTI (d), proposed SuperDTI+ (e) with
additional k-space reduction (l), respectively, and the corresponding difference
map (g-k) with 6 DWIs. The model-fitted tractography from 90 DWIs (d, k, r) is
also shown as a reference.