Alberto De Luca1,2, Suheyla Cetin Karayumak3, Alexander Leemans2, Yogesh Rathi3, Stephan Swinnen4,5, Jolien Gooijers4,5, Amanda Clauwaert4,5, Roald Bahr6, Stian Bahr Sandmo6, Nir Sochen7,8, David Kaufmann9, Marc Muehlmann10, Geert-Jan Biessels1, Inga K Koerte3,11, and Ofer Pasternak3
1Department of Neurology, University Medical Center Utrecht, Utrecht, Netherlands, 2PROVIDI Lab, Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 3Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States, 4Movement Control and Neuroplasticity Research Group, KU Leuven, Leuven, Belgium, 5Brain Institute, KU Leuven, Leuven, Belgium, 6Oslo Sports Trauma Research Center, Norwegian School of Sport Sciences, Oslo, Norway, 7Department of Applied Mathematics, Tel Aviv University, Tel Aviv, Israel, 8Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 9Department of Radiology, Charite University Hospital, Berlin, Germany, 10Department of Radiology, LMU Munich, Munich, Germany, 11cBRAIN, Department of Child and Adolescent Psychiatry, LMU Munich, Munich, Germany
1Department of Neurology, University Medical Center Utrecht, Utrecht, Netherlands, 2PROVIDI Lab, Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 3Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States, 4Movement Control and Neuroplasticity Research Group, KU Leuven, Leuven, Belgium, 5Brain Institute, KU Leuven, Leuven, Belgium, 6Oslo Sports Trauma Research Center, Norwegian School of Sport Sciences, Oslo, Norway, 7Department of Applied Mathematics, Tel Aviv University, Tel Aviv, Israel, 8Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 9Department of Radiology, Charite University Hospital, Berlin, Germany, 10Department of Radiology, LMU Munich, Munich, Germany, 11cBRAIN, Department of Child and Adolescent Psychiatry, LMU Munich, Munich, Germany
The rotation invariant spherical harmonics (RISH) method allows to remove cross-site differences in diffusion kurtosis imaging (DKI) metrics while retaining longitudinal effects, posing a foundation for the application of DKI in large multi-site studies.
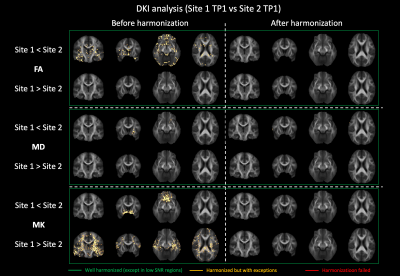
Figure 2: Voxel-wise comparison (t-test) of FA, MD and MK between Site 1 and Site 2 before and after harmonization. Significantly different voxels are shown as yellow overlay. After harmonization, all significant differences were removed.
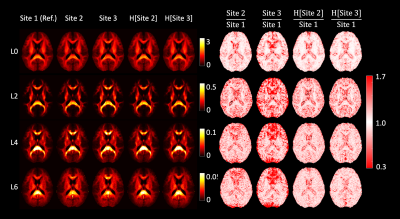
Figure 1: Harmonization of data at b = 2500 s/mm2. The left half of the image shows the RISH features of all three sites before and after harmonization (H[]). The right side shows the voxel-wise scaling between the RISH features of the harmonized site and the reference site. Before harmonization, large scaling can be observed for L0 in periventricular regions, and in L2/L4/L6 in frontal and occipital regions. After harmonization, the cross-site scaling is close to 1.