Zi Wang1, Yihui Huang1, Zhangren Tu2, Di Guo2, Vladislav Orekhov3, and Xiaobo Qu1
1Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden
1Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden
The proof-of-concept
of the significance of merging the optimization with deep learning, for reliable,
high-quality, and ultra-fast accelerated NMR spectroscopy, and provides a
relatively explicit understanding of the complex mapping in deep learning.
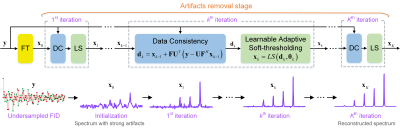
Figure 1. MoDern: The proposed Model-inspired Deep Learning framework for NMR
spectra reconstruction. The recursive MoDern framework that alternates between
the data consistency (DC), which is same to Eq. (1a), and the learnable
adaptive soft-thresholding (LS) inspired by Eq. (1b). With the increase of
iterations, artifacts are gradually removed, and finally a high-quality
reconstructed spectrum can be obtained. Note: “FT” is the Fourier transform. A
data consistency followed by a learnable adaptive soft-thresholding constitutes
an iteration.
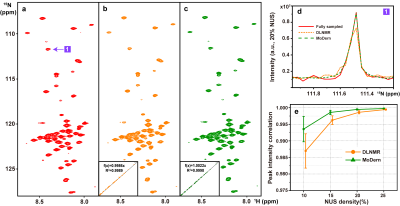
Figure 2. 2D 1H-15N
HSQC spectrum of the cytosolic domain of CD79b protein. (a) The fully sampled
spectrum. (b)-(c) are reconstructed spectra using DLNMR and MoDern from 20%
data, respectively. (d) is zoomed out 1D 15N traces. (e) is the
peak intensity correlation obtained with two methods under different NUS
levels. The insets of (b)-(c) show the corresponding peak intensity correlation
between fully sampled spectrum and reconstructed spectrum. Note: The average and standard deviations of
correlations in (e) are computed over 100 NUS trials.