Kimberly Chan1 and Anke Henning1,2
1The University of Texas Southwestern, Dallas, TX, United States, 2Max Planck Institute for Biological Cybernetics, Tübingen, Germany
1The University of Texas Southwestern, Dallas, TX, United States, 2Max Planck Institute for Biological Cybernetics, Tübingen, Germany
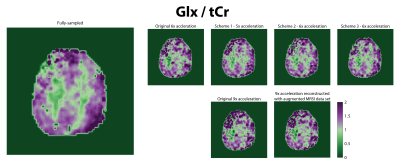
Neural network reconstruction of new CAIPIRINHA-based variable-density k-space undersampling schemes and an approach to train the neural networks by augmenting the MRSI data with the non-water suppressed is introduced and evaluated. Both are shown here to reduce lipid artifacts in MRSI.
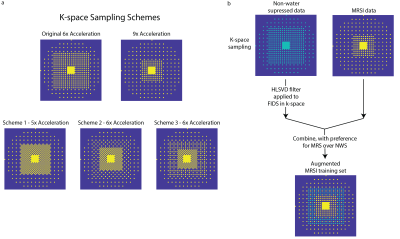
1. (a) K-space undersampling schemes for the originally proposed 6x and 9x-acceleration (first row) as well as the newly proposed variable-density CAIPIRINHA schemes for 5x and 6x acceleration. (b) Schematic for augmenting the MRSI data with water-removed NWS data to provide more k-space points as well as a larger k-space region for training the neural networks based off the lipid behavior.