Kelley M. Swanberg1, Martin Gajdošík1, Karl Landheer1, and Christoph Juchem1,2
1Biomedical Engineering, Columbia University School of Engineering and Applied Science, New York, NY, United States, 2Radiology, Columbia University Medical Center, New York, NY, United States
1Biomedical Engineering, Columbia University School of Engineering and Applied Science, New York, NY, United States, 2Radiology, Columbia University Medical Center, New York, NY, United States
Here we treat spectral baselines as piecewise polynomial shapes akin to metabolite basis
functions to show that amplitude Cramér-Rao Lower Bounds (CRLB) can under some
circumstances offer precision estimates on baseline parameters themselves as well as increase metabolite CRLB accuracy.
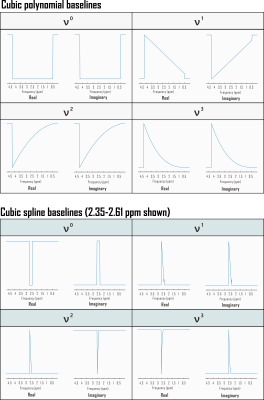
Fig. 1. Partial derivatives of linear combination model with respect to complex baseline shapes for Fisher information matrix calculation. Shown here are the Fourier transforms of example real and imaginary polynomial and spline baseline components incorporated into the Fisher information matrix used to estimate Cramér-Rao Lower Bounds (CRLB) for linear combination model fits to simulated in vivo sLASER (TE 20.1 ms) metabolite proton spectra. Each shape is scaled by its corresponding polynomial coefficient for direct calculation of relative CRLBs. ppm: parts per million.
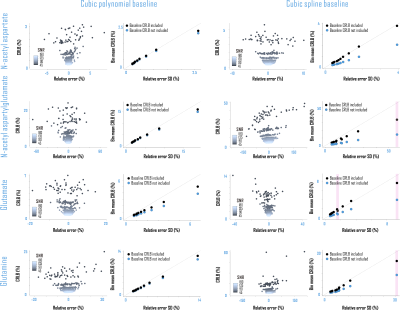
Fig. 5. Baseline Cramér-Rao Lower Bounds (CRLB) improved metabolite CRLB accuracy. Calculating amplitude CRLBs for polynomial or spline baselines directly from the Fisher information matrix improved
correspondence between metabolite amplitude CRLBs and parameter estimate standard deviations (S.D.) by SNR (before=blue; after=black). Correspondent
with Fig. 4, non-normal distributions (pink), for which S.D. may not be an appropriate measure of parameter variability, were observed more often for fits using spline than polynomial
baselines. ppm: parts per million.