HyeongHun Lee1 and Hyeonjin Kim1,2
1Department of Biomedical Sciences, Seoul National University, Seoul, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of
1Department of Biomedical Sciences, Seoul National University, Seoul, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of
The
proposed Bayesian deep learning-based approach in 1H-MRS of the
brain provides both metabolite content and corresponding uncertainty, and
therefore can be advantageous over the standard convolutional neural networks approaches in
consideration of clinical application.
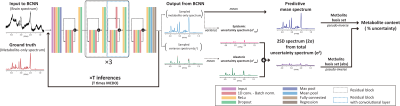
Figure 1. The training of the BCNN and the estimation of metabolite
content and corresponding uncertainty. The metabolite content is estimated by multiple
regression using the metabolite basis set and the predictive mean spectrum that
is the mean spectrum over the BCNN-predicted metabolite-only spectra (number of
spectra = T (number of inferences)). The uncertainty in metabolite content is
estimated by multiple regression using the 2SD spectrum. In this case, the
metabolite basis set is used in absolute mode in accordance with the
2SD-spectrum.
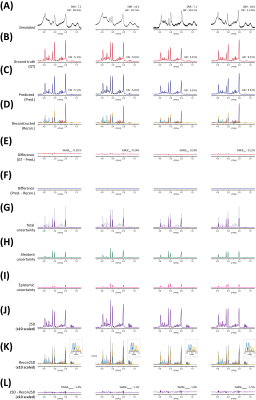
Figure 2. (A) 4 representative simulated
spectra (BCNN input). (B) Ground
truth (GT) spectra. (C) BCNN-predicted
spectra. (D) Reconstructed spectra using the estimated metabolite content and metabolite
bases. (E) Difference spectra (GT – Predicted). (F) Difference spectra (Predicted – reconstructed). (G) Total uncertainty
spectra (= (H) aleatoric + (I) epistemic uncertainty) (BCNN-predicted
spectra shown in dotted line). (J)
2SD spectra. (K) Reconstructed 2SD
spectra using the estimated uncertainty and metabolite bases. (L) Difference spectra (2SD – reconstructed 2SD).
