Neha Koonjoo1,2,3, Adam Berrington4, Bo Zhu2,3,5, Uzay E Emir6,7, and Matthew S Rosen2,3,5
1Department of Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Department of Physics, Harvard University, Cambridge, MA, United States, 4Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom, 5Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 6School of Health Sciences, Purdue University, West Lafayette, IN, United States, 7Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
1Department of Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Department of Physics, Harvard University, Cambridge, MA, United States, 4Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom, 5Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 6School of Health Sciences, Purdue University, West Lafayette, IN, United States, 7Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
A deep neural network based on the AUTOMAP formalism to reconstruct metabolic cycle FIDs into spectral domain. Density matrix formalism was used to generate up/downfields of 1H FIDs of 27 metabolites. The proposed strategy was validated on noisy simulated FIDs and an in vivo cerebellum 3T data.
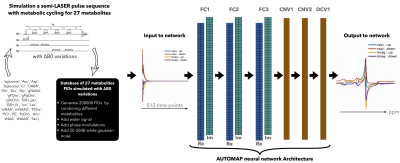
Figure 1a: Proposed strategy to learn the entire end-to-end inverse function and low dimensional representations of spectroscopic signals – (from left to right) Building the training database: The 27 metabolites FIDs were simulated with a sLASER pulse sequence with metabolic cycling and B0 variations. The simulated metabolites FIDs were used to create the training database by combining metabolites, water signal, phase modulations and 20-50dB of gaussian noise. The input/output to the network was the real and imaginary component of metabolic cycle FIDs/spectra (see Methods).
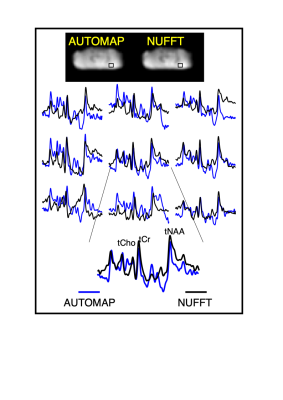
Figure 3: In vivo cerebellum MRSI reconstruction with AUTOMAP. (from top to bottom) The AUTOMAP and NUFFT reconstructed summed image from the 1st time point are shown. The metabolite-only spectra with a range from 0.5 to 4.2 ppm from the 9 different voxels (represented by the black box in the images) are displayed in blue for AUTOMAP and in black for NUFFT. At the bottom an enlarged spectra with the corresponding metabolite peaks (total Choline (tCho), NAA (tNAA) and Creatine (tCr) is represented.
