Milena Capiglioni1,2,3, Analía Zwick1, Pablo Jimenez1, and Gonzalo Álvarez1,2
1Laboratorio de Espectroscopía e Imágenes por RMN, Departamento de Física Médica, Centro Atómico Bariloche - CNEA - CONICET, Bariloche, Argentina, 2Instituto Balseiro, CNEA, Universidad Nacional de Cuyo, Bariloche, Argentina, 3Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, University Hospital of Bern, University of Bern, Bern, Switzerland
1Laboratorio de Espectroscopía e Imágenes por RMN, Departamento de Física Médica, Centro Atómico Bariloche - CNEA - CONICET, Bariloche, Argentina, 2Instituto Balseiro, CNEA, Universidad Nacional de Cuyo, Bariloche, Argentina, 3Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, University Hospital of Bern, University of Bern, Bern, Switzerland
We provide a novel conceptual tool for designing microstructure-size filters with Magnetic Resonance Imaging (MRI) control techniques by selectively probing nuclear-spin dephasing induced by water-molecule diffusion within the tissue-compartments.
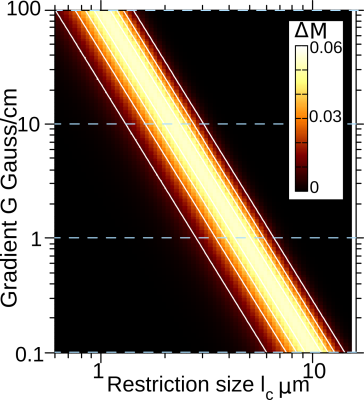
Fig. 3: NOGSE contrast $$$\Delta M$$$as a function of the restriction length $$$l_c$$$ and the gradient strength $$$G$$$, considering $$$N = 8$$$ and a constant value for $$$T_EG$$$ by properly choosing $$$T_E$$$ for each gradient. The considered dynamic range for the gradient strength $$$G$$$ is achievable with current technologies.
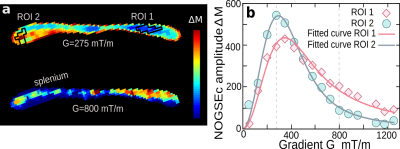
Fig. 4: (a) Two images based on NOGSEc of the Corpus Callosum of an ex-vivo mouse brain for two gradient strengths. For both images $$$N = 2$$$ and $$$T_E = 21.5$$$ ms. (b) Average NOGSEc signal (symbols) as a function of the gradient strength for the ROIs marked in (a). The solid-lines is the fitting of our theoretical model for a log-normal distribution. The fitted parameters are the median $$$1.34 ± 0.04$$$ and $$$2.22 ± 0.02$$$ μm and geometric standard deviation $$$1.82 ± 0.02$$$ and $$$1.82 ± 0.02$$$ μm for ROI 1 and ROI 2 respectively. We considered $$$D_0 = 0.7 μ$$$m$$$^2/$$$ms.
