Yuze Li1, Huijun Chen1, Haikun Qi2, Zhangxuan Hu3, Zhensen Chen1, Runyu Yang1, Huiyu Qiao1, Jie Sun4, Tao Wang5, Xihai Zhao1, Hua Guo1, and Huijun Chen1
1Center for Biomedical Imaging Research, Medical School, Tsinghua University, Beijing, China, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3GE Healthcare, Beijing, China, 4Vascular Imaging Lab and BioMolecular Imaging Center, Department of Radiology, University of Washington, Seattle, Seattle, WA, United States, 5Department of Neurology, Peking University Third Hospital, Beijing, China
1Center for Biomedical Imaging Research, Medical School, Tsinghua University, Beijing, China, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3GE Healthcare, Beijing, China, 4Vascular Imaging Lab and BioMolecular Imaging Center, Department of Radiology, University of Washington, Seattle, Seattle, WA, United States, 5Department of Neurology, Peking University Third Hospital, Beijing, China
A
Deep learning enhAnced T1 parameter mappIng and recoNstruction framework using
spatial-Temporal and phYsical constraint (DAINTY) was proposed. DAINTY imposed
low rank, sparsity and physical constraints to generate good quality T1
weighted images and T1 maps.
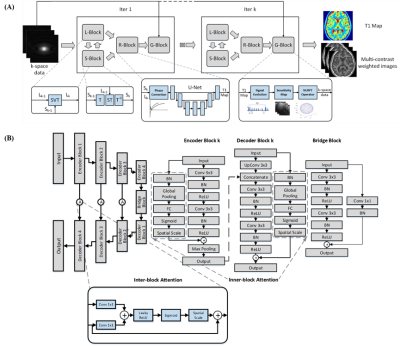
Figure 1. The illustration of DAINTY framework. (A) The under-sampled
k-space data are first processed by L-Block and S-Block to impose the low rank and
sparsity constraint. Then they are processed through R-Block to generate
refined T1 maps. The physical model G-Block is used to transform T1 map back to T1 weighted images to further improve the reconstruction quality. After k iterations the clear T1 map and high-quality T1
weighted images can be both obtained. (B) The structure of DenseAttention UNet.
The number of feature maps for encoder and decoder
block 1-4 are 256, 128, 64, and 32.
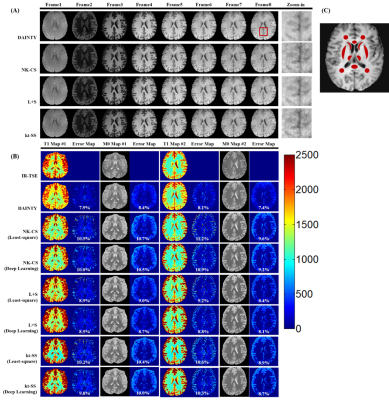
Figure 3. MR images of in-vivo human
brain. (A) T1 weighted images from GOAL-SNAP sequence using DAINTY, NK-CS,
kt-SS and L+S method; (B) Reconstructed T1 and M0 maps and error maps from
GOAL-SNAP sequence using DAINTY, NK-CS, kt-SS and L+S methods with least-square
fitting and direct deep learning mapping. T1 and M0 maps from IR-TSE are as the
reference.