Fan Lam1,2, Hoby Hetherington3, and Jullie Pan3
1Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois Urbana-Champaign, Urbana, IL, United States, 3Radiology, University of Pittsburgh, Pittsburgh, PA, United States
1Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois Urbana-Champaign, Urbana, IL, United States, 3Radiology, University of Pittsburgh, Pittsburgh, PA, United States
We present a new method for rapid 7T MRSI of the brain that uses subspace imaging to improve rosette MRSI (RSI). In vivo results showed substantial improvement over the standard RSI reconstruction and highlight the potentials of integrating acquisition and processing for ultrahigh-field MRSI.
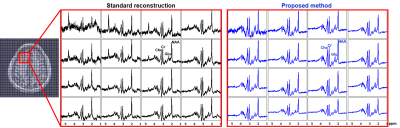
Spatially-localized spectra for a 4x4 voxel array from an anterior brain region are compared, with results produced by the standard reconstruction (see texts) and the proposed method (the blue curves). The location of the region-of-interest is indicated by the red box in the anatomical image on the left with overlaid voxel grids. Significant reduction in lipid leakage and SNR improvement can be observed for the spectra from the proposed method, especially for the edge voxels. Spectra are shown in real parts.
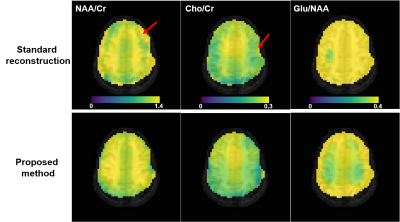
Comparison of NAA/Cr (column 1), Cho/Cr (column 2), Glu/NAA (column 3) maps from the standard reconstruction and the proposed method for one subject. As can be seen, the metabolite ratio maps produced by the proposed method have less artifacts (examples indicated by the red arrows) and better gray/white matter contrast (e.g., the Glu/NAA maps). The color scales were the same for the same column. The metabolite levels were obtained using spectral integration for this preliminary study, and no relaxation correction was performed.
