Peter Adany1, In-Young Choi1,2, and Phil Lee1,3
1Hoglund Biomedical Imaging Center, University of Kansas Medical Center, Kansas City, KS, United States, 2Department of Neurology, University of Kansas Medical Center, Kansas City, KS, United States, 3Department of Radiology, University of Kansas Medical Center, Kansas City, KS, United States
1Hoglund Biomedical Imaging Center, University of Kansas Medical Center, Kansas City, KS, United States, 2Department of Neurology, University of Kansas Medical Center, Kansas City, KS, United States, 3Department of Radiology, University of Kansas Medical Center, Kansas City, KS, United States
Alignment optimization at 10 iterations/sec improves lipid signal removal and better quantitation in
full-field of view MRSI metabolic imaging of the brain.
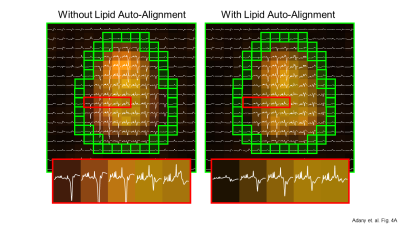
Figure 4A.
MRSI spectra from a subject
with some misalignment between MRI and MRSI. (A) The approximate location of
the lipid mask indicated with green voxels. Lipid signal strength is indicated
by an orange color map. Without lipid auto-alignment, greater and asymmetric residual
lipid signal are clearly visible (4A left). After lipid auto-alignment, the
lipid removal is highly symmetric and the lipid residuals are smaller (4A
right). The estimated displacement was 4.9 mm and 2.5 mm in X and Y directions,
respectively. Spectral range is [0, 4.2] ppm.
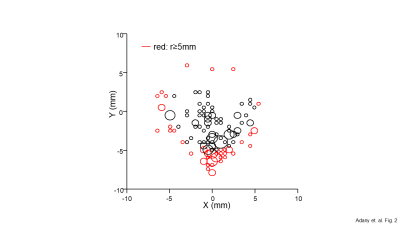
Figure 2. Results of spatial auto-alignment in 141
scans. Auto-alignment was performed on multiple scans of 73 healthy
subjects. Red circles indicate positions outside a 5mm boundary (defined as
Large Displacement). Results showed that coronal head movement was greater than
sagittal during scan sessions, consistent with positioning of the
head within the RF coil in supine position. The alignment precision exceeded the MRSI nominal voxel size of 20x20x25 mm3. Multi-scale
spatial alignment used 7x7 search grids
(40.0mm, 13.5mm, 4.5mm, and 1.5mm) to efficiently locate the minimum.
