Marcia Sahaya Louis1,2, Eduardo Coello2, Huijun Liao2, Ajay Joshi1, and Alexander P Lin2
1Boston University, Boston, MA, United States, 2Brigham and Women's hospital, Boston, MA, United States
1Boston University, Boston, MA, United States, 2Brigham and Women's hospital, Boston, MA, United States
We propose a deep convolutional autoencoder model with feature fusion for removal of water resonance from unsuppressed 97 ms water spectrum and reconstructs an intact metabolite spectrum.
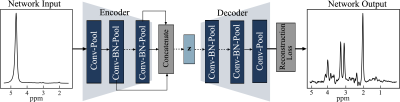
Architecture of Autoencoder with Feature Fusion. The encoder and decoder each have eight convolutional (conv) layers with pooling and batch normalization (BN). Each conv layer had a kernel size of 9 and 16 filters and one fully connected layer with 1000 hidden units. The feature maps of all the layers in the encoder are concatenated using maximum pooling and passed to the latent vector (Z). Z is a fully connected layer with 128 hidden units with a linear activation function. All the layers in the model have a ReLU activation function, except the last layer has a tanh activation function.
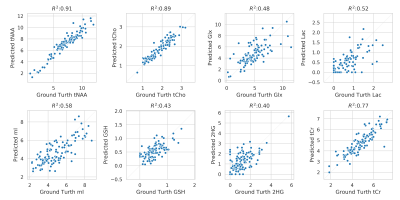
Correlation of metabolite concentration obtained using LCModel, for AE predicted and water suppressed spectra for the in-vivo test dataset. The x-axis shows the absolute concentration values estimated from water suppressed spectra and y-axis shows the corresponding AE predicted spectra. Overall, there is a strong correlation between metabolite for tNAA, tCho, tCr and Glx, Lac, GSH, mI and 2HG has a decent correlation. The clustering pattern seen in other metabolites can be caused by reconstruction error and change in the degree of freedom of LCModel fit between the spectra.
