Daniel ZL Kor1, Saad Jbabdi1, Jeroen Mollink1, Istvan N Huszar1, Menuka Pallebage- Gamarallage2, Adele Smart2, Connor Scott2, Olaf Ansorge2, Amy FD Howard1, and Karla L Miller1
1Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 2Neuropathology, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
1Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 2Neuropathology, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
Here, we describe an end-to-end pipeline for the extraction of a histological metric from IHC stains to quantify a microstructural feature. We compare the pipeline's reproducibility and robustness to histology artefacts, relative to manual MRI-histology analyses.
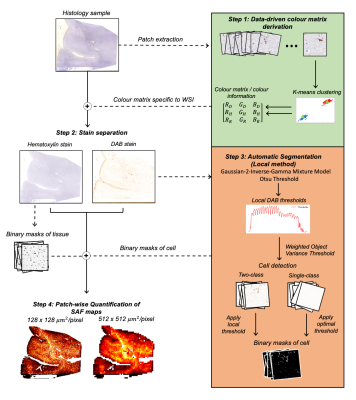
Fig. 1: A robust, automatic pipeline to quantify the stained area fraction (SAF) from histology slides as highlighted in the 4 steps. Input RGB slides are processed to produce SAF maps at variable resolution. We aim to correlate SAF maps at MRI-scale resolution (512x512 µm2/pixel) to MRI measures.
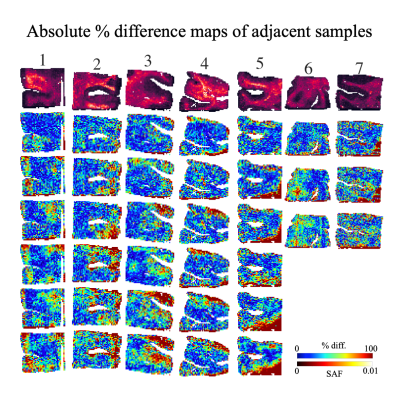
Fig. 3: An example SAF map (top row) and the absolute percent difference maps for all within-subject pairwise comparisons between adjacent slides with local thresholding (i.e. proposed pipeline). Each column shows a different subject. 3 adjacent slides produce 3 pairwise comparisons (subjects 6,7), while 4 adjacent slides produce 6 pairwise comparisons (other subjects). For almost all subjects, the highest percentage difference is found on the edges of the tissue, implying possible misalignment after co-registration or reduced robustness to tissue edge artefacts.
