Chongxue Bie1,2,3, Yang Zhou4, Peter C. M. van Zijl1,2, Jiadi Xu1,2, Ramon C. Sun5, Matthew S. Gentry66, and Nirbhay N. Yadav1,2
1The Russell H. Morgan Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Information Science and Technology, Northwest University, Xi'an, China, 4Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 5Department of Neuroscience, University of Kentucky, Lexington, KY, United States, 6Department of Molecular and Cellular Biochemistry, University of Kentucky, Lexington, KY, United States
1The Russell H. Morgan Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Information Science and Technology, Northwest University, Xi'an, China, 4Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 5Department of Neuroscience, University of Kentucky, Lexington, KY, United States, 6Department of Molecular and Cellular Biochemistry, University of Kentucky, Lexington, KY, United States
glycoNOE MRI was applied to study brain glycogen accumulation in a mouse model of Lafora disease. In-vivo glycoNOE maps resemble histology, suggesting that this approach has potential for disease diagnosis, monitoring progression, and treatment response.
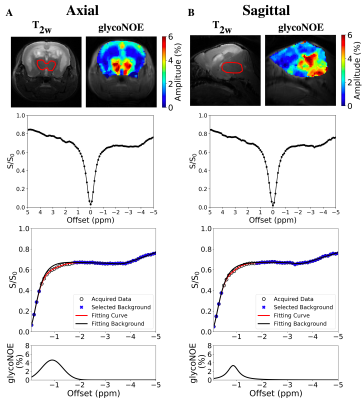
Figure 1. glycoNOE MRI on brain of the Lafora disease mouse model. (A) Axial and (B) sagittal slices. Data were acquired with an RF saturation power of B1 =0.7 μT. ROIs are indicated in T2w images by a red line. The Z-spectrum was fitted with Lorentzian and Gaussian hybrid line-shape, by assuming multi-Lorentzian functions for background (blue mark) and Voigt function for glycoNOE (shown in the bottom row). The glycoNOE map was estimated from the amplitude of the corresponding Voigt line-shape.
