Yuki Asada1, Luke Xie2, Skander Jemaa3, Kai H. Barck2, Tracy Yuen4, Richard A.D. Carano3, and Gregory Z. Ferl1
1Pharmacokinetics & Pharmacodynamics, Genentech, Inc., South San Francisco, CA, United States, 2Biomedical Imaging, Genentech, Inc., South San Francisco, CA, United States, 3PHC Data Science Imaging, Genentech, Inc., South San Francisco, CA, United States, 4Neuroscience, Genentech, Inc., South San Francisco, CA, United States
1Pharmacokinetics & Pharmacodynamics, Genentech, Inc., South San Francisco, CA, United States, 2Biomedical Imaging, Genentech, Inc., South San Francisco, CA, United States, 3PHC Data Science Imaging, Genentech, Inc., South San Francisco, CA, United States, 4Neuroscience, Genentech, Inc., South San Francisco, CA, United States
A fully convolutional U-Net neural network for 3D images is trained to automatically detect and segment brain lesions in MRI scans of the cuprizone mouse model for multiple sclerosis, where impact of image preprocessing and augmentation steps on U-Net performance is evaluated.
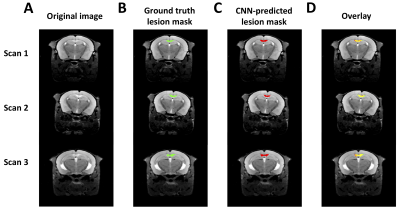
Figure 1. Three representative hold-out test scans. A) The transverse slice approximately through center of the lesion is shown representative scans, along with B) Ground truth and C) CNN-predicted ROIs, using the trained model indicated in Table 1, row 8. D) An overlay of the 2 ROIs is shown, with overlapping area indicated in yellow.
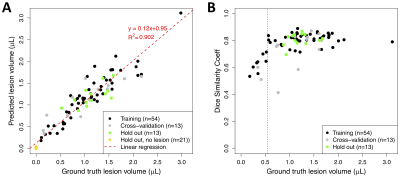
Figure 4. Per-animal predicted volumes and dice similarity coefficients. A) Distribution of DSC achieved during training (network in Figure 2, row 8) vs. volume (units are number of voxels) of ground truth ROIs. B) Predicted ROI volume vs. ground truth volume (units are number of voxels). Prediction is performed using trained network shown in Figure 2, row 8.
