Dipika Sikka1,2, Noah Igra3,4, Sabrina Gjerswold-Sellec1, Cynthia Gao5, Ed Wu6, and Jia Guo7
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2VantAI, New York, NY, United States, 3Department of Applied Mathematics, Columbia University, New York, NY, United States, 4Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel, 5Department of Computer Science, Columbia University, New York, NY, United States, 6Department of Electrical and Electronic Engineering, The University of Hong Kong, Pok Fu Lam, Hong Kong, China, 7Department of Psychiatry, Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2VantAI, New York, NY, United States, 3Department of Applied Mathematics, Columbia University, New York, NY, United States, 4Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel, 5Department of Computer Science, Columbia University, New York, NY, United States, 6Department of Electrical and Electronic Engineering, The University of Hong Kong, Pok Fu Lam, Hong Kong, China, 7Department of Psychiatry, Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
Completely complex U-Net architectures, developed with novel application of Attention, Pooling, and Residual components, demonstrate significant utility and potential in MR k-space signal processing.
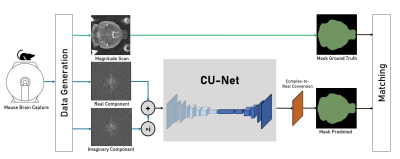
Figure 1. Complex domain pipeline for mouse-brain extraction. Raw data is used to generate real and imaginary components for k-space data, which was used to train the complex Residual Attention U-Net network. Model output was compared to ground truth mouse-brain masks.
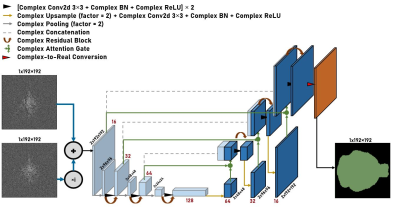
Figure 2. Sample complex network Residual Attention U-Net architecture. This network shows the architecture of one of the complex networks trained for the mouse-brain extraction task. The network consists of 4 encoding layers and 4 decoding layers. Spatial dimension decreases by 2 and channel dimension increases by 2 as the data propagates through the encoding layers while the reverse happens along the decoding layers. A similar structure is used for networks with 3, 5, and 6 layers.
