Kaiyue Tao1, Li Chen2, Niranjan Balu3, Gador Canton3, Wenjin Liu3, Thomas S. Hatsukami4, and Chun Yuan3
1University of Science and Technology of China, Hefei, China, 2Department of Electrical and Computer Engineering, University of Washington, Seattle, WA, United States, 3Department of Radiology, University of Washington, Seattle, WA, United States, 4Department of Surgery, University of Washington, Seattle, WA, United States
1University of Science and Technology of China, Hefei, China, 2Department of Electrical and Computer Engineering, University of Washington, Seattle, WA, United States, 3Department of Radiology, University of Washington, Seattle, WA, United States, 4Department of Surgery, University of Washington, Seattle, WA, United States
We explored vessel wall imaging information hidden in MRI images from the OAI dataset. We designed a metric learning network combined with an episodic training method to overcome the problem of limited annotations, and demonstrated its ability to learn a meaningful feature space.
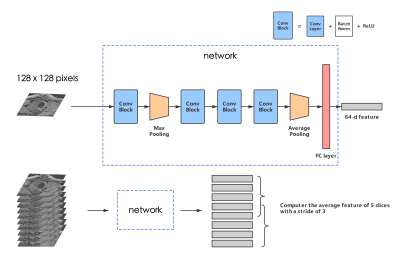
Figure 2 - The structure of our network. A Conv Block consists of a CNN layer, a batch normalization layer, and an active function ReLU. The kernel size of each convolution layer: 7*7, 3*3, 3*3, 3*3. A feature map with the size of (128, 7, 7) is outputted from the last Conv Block, and will then be reshaped into (128, 1) after max pooling. The number of total param of the network is 129600.
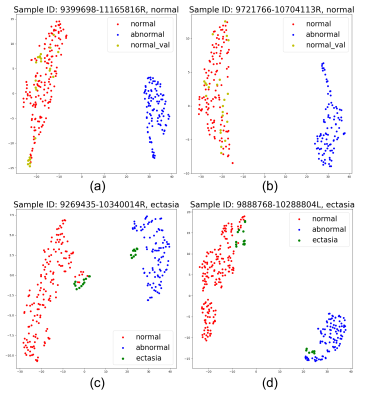
Figure 4 - Feature map of validation samples with normal/abnormal clusters in 2D space using t-SNE. Each dot represents an average feature of 5 slices in MRI scans, as described in the "inference" part. The colors (red for normal, blue for abnormal (aneurysm), green for ectasia, and yellow for normal validation samples) are painted to show the type of each dot. (a) and (b) show 2 normal validation samples, and (c) and (d) show 2 ectasia samples. The clusters' position can vary due to the t-SNE algorithm, as shown in (c) and (d), but the distance between features will be truly presented.