Ina Hanninger1, Eylem Levelt2,3, Jennifer J Rayner2, Christopher T Rodgers2,4, Stefan Neubauer2, Vicente Grau1, Oliver J Rider2, and Ladislav Valkovic2,5
1Oxford Institute of Biomedical Engineering, Oxford, United Kingdom, 2Radcliffe Department of Medicine, University of Oxford Centre for Clinical Magnetic Resonance Research, Oxford, United Kingdom, 3University of Leeds, Leeds, United Kingdom, 4Wolfson Brain Imaging Centre, Cambridge, United Kingdom, 5Slovak Academy of Sciences, Institute of Measurement Science, Bratislava, Slovakia
1Oxford Institute of Biomedical Engineering, Oxford, United Kingdom, 2Radcliffe Department of Medicine, University of Oxford Centre for Clinical Magnetic Resonance Research, Oxford, United Kingdom, 3University of Leeds, Leeds, United Kingdom, 4Wolfson Brain Imaging Centre, Cambridge, United Kingdom, 5Slovak Academy of Sciences, Institute of Measurement Science, Bratislava, Slovakia
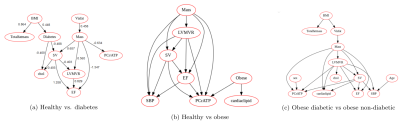
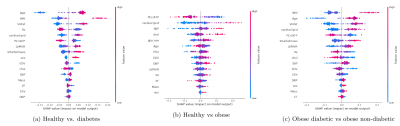
Random Forest classifiers and Bayesian networks applied to MR spectroscopy measures suggests a high predictive impact of cardiac metabolism in classifying diabetic and obese patients, further implying a causal mechanism with visceral fat, concentric LV remodeling and energy impairment