Hengda He1, Linbi Hong1, and Paul Sajda1,2,3,4
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Electrical Engineering, Columbia University, New York, NY, United States, 3Department of Radiology, Columbia University, New York, NY, United States, 4Data Science Institute, Columbia University, New York, NY, United States
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Electrical Engineering, Columbia University, New York, NY, United States, 3Department of Radiology, Columbia University, New York, NY, United States, 4Data Science Institute, Columbia University, New York, NY, United States
We propose a locus coeruleus localization and BOLD activity extraction method, which shows significant correlation with trial-to-trial variability in baseline pupil diameter.
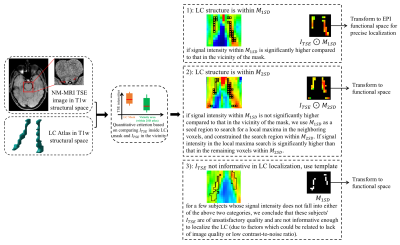
Figure 1. LC localization using a predefined LC atlas and the TSE image of each subject. In T1w structural space, we use a criterion (Student’s t-test) to determine a coarse LC location. Then, either TSE intensities in the LC mask or an LC atlas is transformed to EPI functional space for a more precise localization (using trilinear interpolation). The result is one of three possible outcomes: 1) we localize the LC structure within M1SD; 2) we localize the LC structure within M2SD; 3) we localize the LC structure with the predefined LC atlas (i.e., without using any information from ITSE).
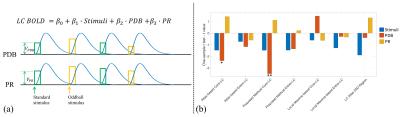
Figure 3. (a) GLM to estimate and test the contributions of PDB and PR to LC BOLD activity (controlling for the variance due to the presence of stimuli). Trial-to-trial variabilities of PDB and PR (VPDB and VPR) were modeled as boxcar functions with the amplitude of each trial modulated by the pupil measurements. The boxcar functions were convolved with a canonical double-gamma hemodynamic response function before fitting into the GLM. (b) Group level statistical analysis in testing regression weights against zero. *p < 0.05; **p < 0.01.