Marianna Inglese1, Shah Islam1, Matthew Grech-Sollars1,2, Giulio Anichini3, James Davies4, Azeem Saleem4,5, Matthew Williams6,7, Kevin S O'Neill3, Adam D Waldman8, and Eric O Aboagye1
1Surgery and Cancer, Imperial College London, London, United Kingdom, 2Imaging, Imperial College London Healthcare NHS Trust, London, United Kingdom, 3Imperial College London Healthcare NHS Trust, London, United Kingdom, 4Invicro Imperial College London, London, United Kingdom, 5Hull York Medical School, Faculty of Health Sciences, University of Hull, Hull, United Kingdom, 6Computational Oncology Group, Department of Surgery and Cancer, Imperial College London, London, United Kingdom, 7Institute for Global Health Innovation, Imperial College London, London, United Kingdom, 8Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
1Surgery and Cancer, Imperial College London, London, United Kingdom, 2Imaging, Imperial College London Healthcare NHS Trust, London, United Kingdom, 3Imperial College London Healthcare NHS Trust, London, United Kingdom, 4Invicro Imperial College London, London, United Kingdom, 5Hull York Medical School, Faculty of Health Sciences, University of Hull, Hull, United Kingdom, 6Computational Oncology Group, Department of Surgery and Cancer, Imperial College London, London, United Kingdom, 7Institute for Global Health Innovation, Imperial College London, London, United Kingdom, 8Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
A strong correlation was found
between 18F-FPIA PET SUV and K1, and between the Ki and the MRI perfusion parameters rCBF and rCBV. By combining PET and MRI, the preliminary
grade predictive vector provided 100% accuracy in tumour grade prediction.
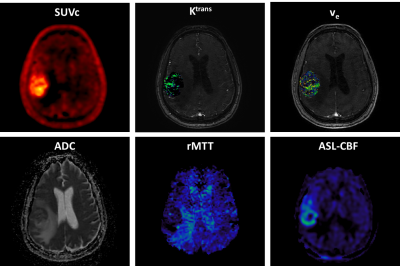
Figure 1: 18F-FPIA
PET/MRI acquisition. Axial
images of a representative patient of the standardised uptake value corrected
by the tracer uptake in the whole blood (SUVc), the contrast agent
plasma/interstitium transfer rate constant Ktrans and the
extracellular extravascular volume fraction ve resulted from the
post-processing of DCE-MRI data, the apparent diffusion coefficient (ADC) map
from DWI-MRI acquisition, the relative contrast agent mean transit time (rMTT)
derived from DSC-MRI and the cerebral blood flow (CBF) derived from the
sequence of ASL.
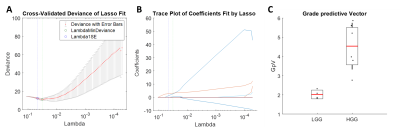
Figure 2: The least absolute shrinkage and selection operator (LASSO)
was applied to extract a composite vector - the most significant tumour grade
predictor - from 25 parameters determined by analysis of simultaneously
acquired PET and MRI data. Five-fold cross-validation was performed to select
lambda minimum to give the minimum cross-validated error (A). The
weighted sum of 3 selected features (B) gave the Grade predictive Vector
GpV, which showed an AUC and accuracy of 1 in the discrimination between low
and high-grade gliomas (C).
