Sana Vaziri1, Adam Autry1, Yaewon Kim1, Hsin-Yu Chen1, Jeremy W Gordon1, Marisa LaFontaine1, Jasmine Graham1, Janine Lupo1, Jennifer Clarke2, Javier Villanueva-Meyer1, Nancy Ann Oberheim Bush3, Duan Xu1, Susan M Chang2, Peder EZ Larson1, Daniel B Vigneron1,4, and Yan Li1
1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Department of Neurological Surgery, University of California, San Francisco, San Francisco, CA, United States, 3Department of Neurology, University of California, San Francisco, San Francisco, CA, United States, 4Department of Bioengineering and Therapeutic Science, University of California, San Francisco, San Francisco, CA, United States
1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Department of Neurological Surgery, University of California, San Francisco, San Francisco, CA, United States, 3Department of Neurology, University of California, San Francisco, San Francisco, CA, United States, 4Department of Bioengineering and Therapeutic Science, University of California, San Francisco, San Francisco, CA, United States
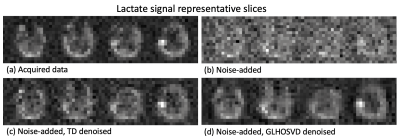
An evaluation of two higher-order singular value decomposition denioising techniques is presented on hyperpolarized [13C]pyruvate metabolic images acquired on patients with glioma. Methods are evaluated using simulations and patient data. After denoising, an increase in SNR is observed.
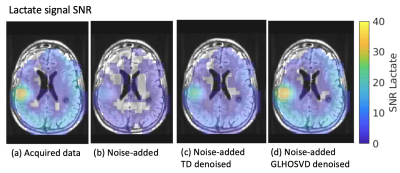
SNR maps for area-under-the-curve (AUC) lactate images (obtained by summing images over all 20 time points) after applying kPL error mask. Both denoising methods (c, d) recover more voxels than the artificially noise-added data (b) and are comparable to the originally acquired data (a). The GLHOSVD-denoised SNR map better agrees with the originally acquired data.