Jan N Rose1, Andrew D Scott2,3, and Denis J Doorly1
1Department of Aeronautics, Imperial College London, London, United Kingdom, 2Cardiovascular Magnetic Resonance unit, Royal Brompton Hospital, London, United Kingdom, 3National Heart and Lung Institute, Imperial College London, London, United Kingdom
1Department of Aeronautics, Imperial College London, London, United Kingdom, 2Cardiovascular Magnetic Resonance unit, Royal Brompton Hospital, London, United Kingdom, 3National Heart and Lung Institute, Imperial College London, London, United Kingdom
We have developed an algorithm to pack cardiomyocytes of arbitrary polygonal shape into groups also of arbitrary shape, known as sheetlets, found in the myocardium. Using the method, we synthesise an imaging voxel replicating histological specimen geometric properties.
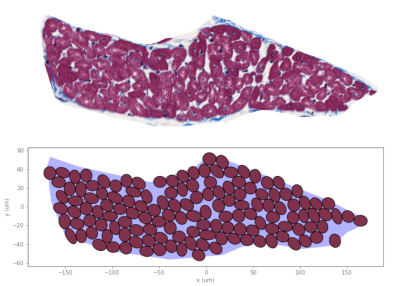
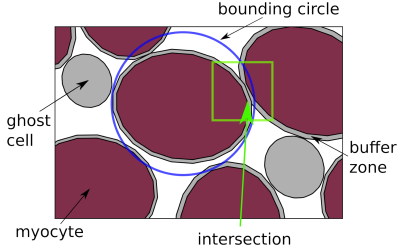
(Top) Sheetlet segmented from histological images. (Bottom) Synthesised sheetlet after completion of the packing algorithm and after removing ghost cells, which leave distinct pockets of ECS between myocytes. The sheetlet outline was used as the target region and myocytes were created to match cell geometries from histology.