Wei Zhang1, Eva M Palacios1, and Pratik Mukherjee1
1UCSF, San Francisco, CA, United States
1UCSF, San Francisco, CA, United States
We introduce deep linear models for
hierarchical fMRI brain connectivity network reconstruction that do not require the manual
hyperparameter tuning, extensive fMRI training data or high-performance
computing infrastructure of nonlinear deep learning.
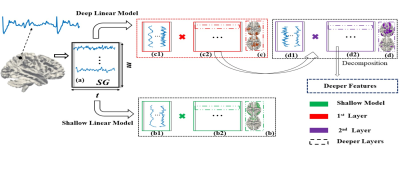
Figure 1. Deep linear model (shown as (c),
(d)) versus shallow linear model (shown as (b)). (a)
SG represents the input fMRI signal matrix, containing the t time
points and m voxels. (c1) and
(d1) represents the weight matrix/dictionary identified via SG of 1st
and 2nd layer, respectively. (c2)
and (d2) represents the feature matrix of 1st and 2nd
layer, respectively. The dashed blue rectangle indicates the deeper features
beyond the 2nd layer.
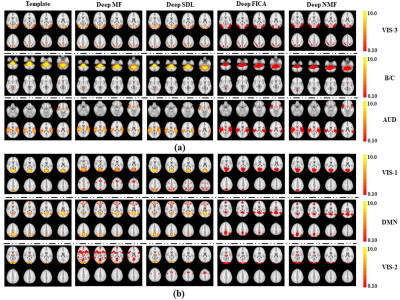
Figure 2. Comparison of six representative 1st
layer networks from all four deep linear models (presented in the second to
fifth column) with the ground truth templates (presented in the first column)
from simulated fMRI data. (a) Three networks
illustrate better intensity matching to the templates by Deep MF and Deep SDL
than by Deep FICA or Deep NMF. (b) Three
networks show better spatial matching to the templates by Deep FICA and Deep
NMF than Deep MF or Deep SDL. Auditory Network: AUD. Brainstem/Cerebellum: B/C. Default Mode Network: DMN. Visual networks: VIS-1, VIS-2, VIS-3.
