Olivia Viessmann1, Divya Varadarajan1, Adrian V Dalca1,2, Bruce Fischl1,2, Michael Bernier1, Lawrence L Wald1,2, and Jonathan R Polimeni1,2
1Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 2Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
1Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 2Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
We provide a proof of concept that surface-based CNNs can predict anatomical and physiological data from fMRI signals. Specifically, we trained CNNs to predict local cortical thickness, cortical orientation to the B0 field and MR angiography data from resting-state timeseries.
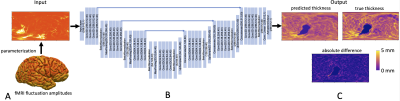
Figure 1: Overview of deep learning framework. A) The surface-based data is parameterized using spherical coordinates to a 2D “image” to serve as CNN input. B) A U-Net architecture is used to learn the relationship to the output data. Outputs are 2D “images” of anatomical or physiological features also parameterized using spherical coordinates. C) Example estimation of cortical thickness.
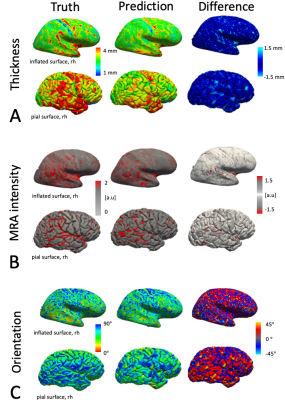
Figure 2: Results of the CNN predictions for an exemplar test subject. True and predicted thickness (A), MRA intensity (B) and cortical orientation angles (C). Feature maps are displayed on the inflated and pial surface. The prediction error varies spatially, but generally the pattern of error is unstructured with some focal regions exhibiting higher predictability than others. Generally, the predicted feature maps appear to be a smoothed version of the ground truth feature maps.
