Bochao Li1, Nam G. Lee1, and Krishna Nayak2
1Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States
1Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Ming Hsieh Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States
We propose a framework for predicting
signal loss in lung parenchyma MRI. Simulations predict that signal loss is
dependent on B0 field strength and insensitive to
respiratory phase. Proximity to alveoli dominates over proximity to the
bronchial tree.
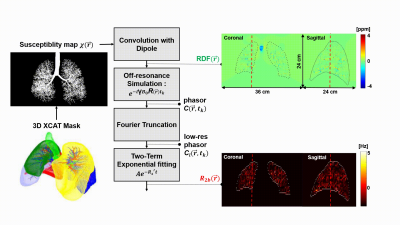
Figure 1: Bronchial
Tree Simulation (R2b) We use a recent lung XCAT phantom to generate a susceptibility
map with the entire bronchial tree. This is used to
compute a high resolution RDF map, that is smoothed using Fourier truncation. Several timepoints are then simulate, and exponential fitting
is used to estimate R2b maps. RDF maps and R2b maps are shown with an overlaid lung outline. Dashed
lines indicate the location of orthogonal sections.
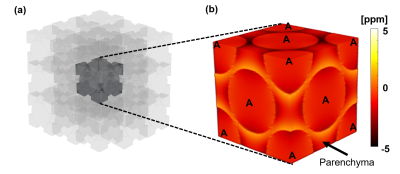
Figure 2: Alveolus
Simulation (R2b). We simulate face-center cubic packing, using (a) a 9x9x9
lattice of fundamental blocks (3x3x3 is illustrated). Each fundamental block represents
0.5x0.5x0.5 mm3
with 5
isotropic resolution. We then simulate the RDF, and extract the (b) central
fundamental block from the RDF map. We exclude air-filled spheres, and use only
parenchyma voxels to simulate dephasing and perform estimation. A = alveolus.
