Wallace Souza Loos1,2, Roberto Souza2,3, Linda Andersen1,2, R. Marc Lebel2,4, and Richard Frayne1,2
1Radiology and Clinical Neuroscience, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 2Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, AB, Canada, 3Electrical and Computer Engineering, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 4General Electric Healthcare, Calgary, AB, Canada
1Radiology and Clinical Neuroscience, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 2Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, AB, Canada, 3Electrical and Computer Engineering, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 4General Electric Healthcare, Calgary, AB, Canada
A deep learning approach was used to estimate a
vascular function from dynamic contrast magnetic images. Our model was able to
generalize well for unseen data and achieved a good overall performance without
requiring manual intervention or major preprocessing steps.
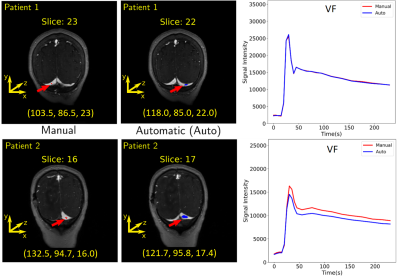
Estimation of the region and
the VF for two patients. The first column shows where the manual region was
drawn. The coordinate of the center of mass (in voxels) is placed below each
image. The second column shows the predicted region. It is possible to observe
that different regions over the transverse sinus can yield similar vascular
functions, as illustrated in the plots of the third column. Plots: Red = manual
VF and blue = predicted VF.
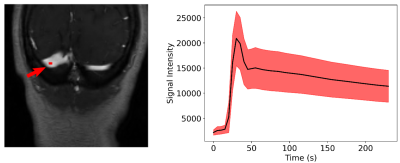
A region of interest is
selected manually over the transverse sinus (left) to estimate the VF (right). To
compute the VF, the region was propagated across the dynamic T1-weighted
images and the average of the intensities of the pixels was computed for each time
point. The mean (black line) and standard deviation (red shaded region) of the 155
VF curves is presented on the right image.
