Tianjia Zhu1,2, Minhui Ouyang1, Nikou Lei3, David Wolk4, Paul Yushkevich5, and Hao Huang1,5
1Department of Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3Department of Physics, University of Washington Seattle, Seattle, WA, United States, 4Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 5Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
1Department of Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3Department of Physics, University of Washington Seattle, Seattle, WA, United States, 4Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 5Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
We
show that a data-driven machine learning approach developed using registered high-resolution
dMRI and histological data of the macaque brain can predict soma and neurite
densities from DTI and DKI metrics.
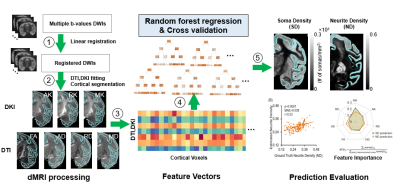
Fig.1 Soma
and neurite densities (SD and ND) estimation from diffusion MRI metrics. Left:
Diffusion weight images registration and DTI, DKI fitting. Cortical
segmentation and overlay on fitted parameter maps. axial kurtosis (AK), radial kurtosis (RK), mean kurtosis (MK),
fractional anisotropy (FA), axial diffusivity (AD), radial diffusivity (RD),
mean diffusivity (MD). Middle: Each voxel in the cortex served as a training
sample in random forest regressor. Right: Estimated SD and ND values. Estimation
evaluation and extraction of feature importance from model.
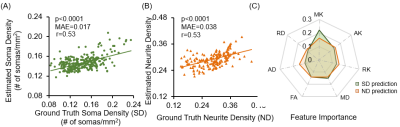
Fig.4 Soma
and neurite densities (SD and ND) estimation results and feature importance. A: Significant linear correlation between ground truth SD and estimated SD on validation set. (P<0.0001, mean absolute error (MAE)=0.017, Pearson
correlation coefficient r=0.53). B: Significant linear correlation between ground
truth ND and estimated ND on validation set. (P<0.0001, MAE= 0.038, Pearson
correlation coefficient r=0.53). Panel C: Feature importance for each metric.
MK ranks firsts for both SD and ND estimation.
