Christopher S Parker1, Thomas Veale2, Martina Bocchetta2, Catherine F Slattery2, Nick Fox2, Jonathan M Schott2, Dave M Cash1,2, and Hui Zhang1
1Centre for Medical Image Computing, Department of Computer Science, UCL, London, United Kingdom, 2Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square, Institute of Neurology, UCL, London, United Kingdom
1Centre for Medical Image Computing, Department of Computer Science, UCL, London, United Kingdom, 2Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square, Institute of Neurology, UCL, London, United Kingdom
Tissue-weighted means utilise the tissue fraction from NODDI to calculate the average microstructural property of the tissue compartments within a region. This provides unbiased estimates of region tissue microstructure when microstructure spatially covaries with tissue fraction.
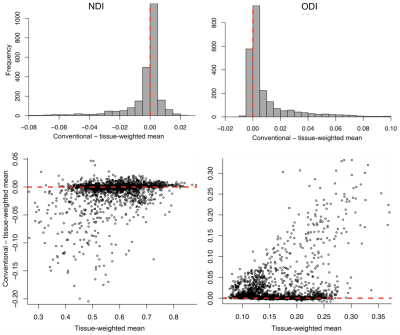
Figure 1. Upper: Histogram of difference between conventional and tissue-weighted means for NDI (left) and ODI (right). The heavy tails of the distributions demonstrate how the conventional mean was in general a biased estimate of the tissue mean, under- and over-estimating NDI and ODI, respectively. Lower: Bland-Altmann style plot of the difference between the conventional and tissue-weighted means across all ROIs and subjects (2448 ROIs). The difference tends to decrease and increase as a function of tissue-weighted mean for NDI and ODI, respectively.
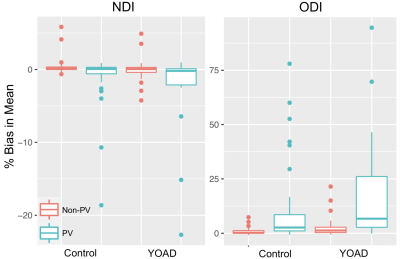
Figure 2. Percentage bias in the ROI between-subject averaged conventional means, for NDI (left) and ODI (right). Each boxplot shows the distribution across ROIs of the percentage difference in the between-subject average conventional and tissue-weighted mean for PV and non-PV ROIs in control and YOAD subjects. NDI tended to be under-estimated, apart from control subjects non-PV ROIs, which were over-estimated. ODI was over-estimated in PV and non-PV ROIs for control and YOAD subjects.