Abdullah S. Bdaiwi1,2, Matthew M. Willmering1, Hui Wang3, and Zackary I. Cleveland1,2,4,5
1Center for Pulmonary Imaging Research, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Biomedical Engineering Department, University of Cincinnati, Cincinnati, OH, United States, 3Philips, Cincinnati, OH, United States, 4Department of Pediatrics, University of Cincinnati, Cincinnati, OH, United States, 5Division of Pulmonary Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
1Center for Pulmonary Imaging Research, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Biomedical Engineering Department, University of Cincinnati, Cincinnati, OH, United States, 3Philips, Cincinnati, OH, United States, 4Department of Pediatrics, University of Cincinnati, Cincinnati, OH, United States, 5Division of Pulmonary Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
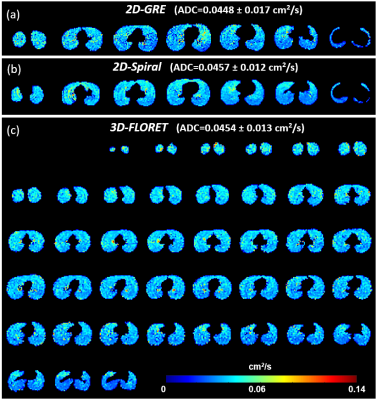
2D and 3D spiral
sequences enable fast acquisition of diffusion-weighted 129Xe data,
enabling full lung coverage at high resolution or with reduced breath holds.
Resulting ADC maps are comparable qualitatively and quantitatively to those
obtained via conventional 2D GRE sequences.
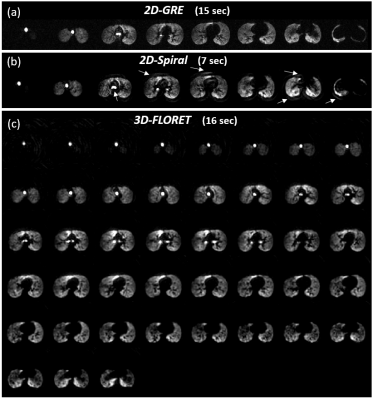
Figure 1: Representative b0
images for (a) 2D-GRE, (b) 2D-spiral and (c) 3D-FLORET sequences.
Spiral artifacts present near the edges of high signal regions are indicated with
arrows. Scan time was 7 s (0.21 ms per voxel) for 2D-spiral and 16 s (0.11 ms
per voxel) for 3D-FLORET compared to 15 s (0.73 ms per voxel) for 2D-GRE.