Paul B Camacho1,2,3, Evan D Anderson3,4, Aaron T Anderson3,5, Hillary Schwarb3, Tracey M Wszalek3, and Brad P Sutton1,2,3
1Neuroscience Program, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Decision Neuroscience Laboratory, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5Stephens Family Clinical Research Institute, Carle Foundation Hospital, Urbana, IL, United States
1Neuroscience Program, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Decision Neuroscience Laboratory, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5Stephens Family Clinical Research Institute, Carle Foundation Hospital, Urbana, IL, United States
We developed a wrapper for
performing resting-state functional connectivity analysis via Singularity
images on high-performance computing systems. This pipeline provides
network-based statistics, data quality, and documentation.
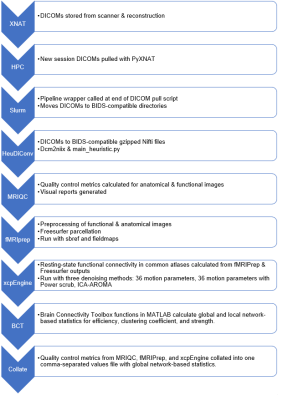
Flow of pipeline from raw DICOMs to end-user entry
into a Research Electronic Data Capture (REDCap) study management database. XNAT: Extensible Neuroimaging Archive Toolkit, HPC: High Performance Computing system, Slurm: Slurm Workload Manager, HeuDiConv: Heuristic-centric DICOM Converter, MRIQC: Magnetic Resonance Imaging Quality Control, fMRIPrep: Function Magnetic Resonance Imaging Preprocessing, xcpEngine: the XCP Engine, BCT: Brain Connectivity Toolbox.
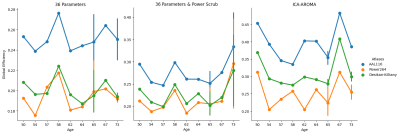
Comparison of global efficiency as
calculated by the Brain Connectivity Toolbox MATLAB functions for two common
structural atlases (Automated Anatomical Labeling 116, Desikan-Killiany) and
the Power 264 functional atlas after three methods of denoising in xcpEngine
(36 motion parameters, 36 motion parameters and Power scrub method, and
ICA-AROMA).
