Gehua Tong1, Sairam Geethanath2, and John Thomas Vaughan, Jr.2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
We present an open-source Python tool, py2jemris, which translates arbitrary Pulseq files for JEMRIS simulation. A dual simulation/acquisition experiment using the same sequence file was performed to demonstrate the pipeline.
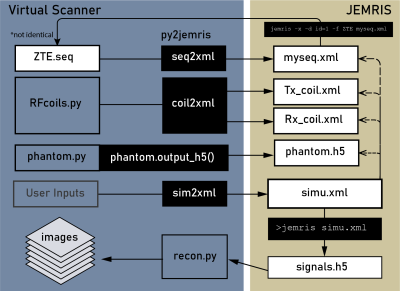
Figure 1: The simulation pipeline enabled by bridging Pulseq to JEMRIS. Sequence, phantom, and B1 map files are converted into JEMRIS format, simulation is performed on the JEMRIS command line, and the resulting data is reconstructed automatically to yield images.
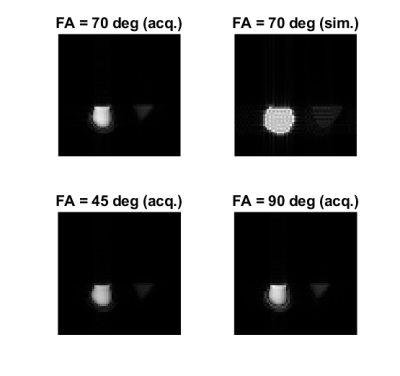
Figure 4: Simulating and acquiring 64 x 64 phantom images from the same sequence file (FOV = 250 mm, slice thickness = 5 mm, TR = 4500 ms, TE = 50 ms). Simulation at FA = 70 degrees was performed while images were acquired with FA = 45, 70, and 90 degrees. At FA = 70 degrees, PSNR = 19.12 dB and SSIM = 0.7531 between the simulated and the acquired images.
