Can Wu1,2 and Qi Peng3
1Department of Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Philips Healthcare, Andover, MA, United States, 3Department of Radiology, Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, NY, United States
1Department of Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Philips Healthcare, Andover, MA, United States, 3Department of Radiology, Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, NY, United States
Deep learning can be used to effectively eliminate synovial fluid from
T1ρ data acquired without fluid suppression, potentially leading to improved
T1ρ quantification accuracy of knee cartilage without adding scan time.
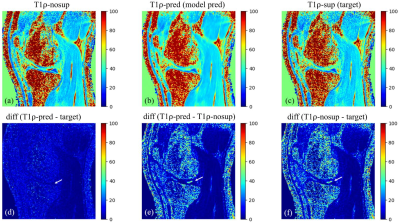
Figure 3.
Example images of T1ρ-nosup (a) and T1ρ-sup (c)
from conventional curve fitting method, along with the T1ρ map predicted by the
deep learning model (b). Synovial fluid (SF) can be easily identified near the
knee cartilage on the T1ρ-nosup image (a), while this was largely suppressed on
images of the T1ρ-sup (c) and the T1ρ-pred (b). Pairwise absolute difference
images (d-f) further illustrate that SF is selectively suppressed (white
arrows) without changing the T1ρ of the cartilage.
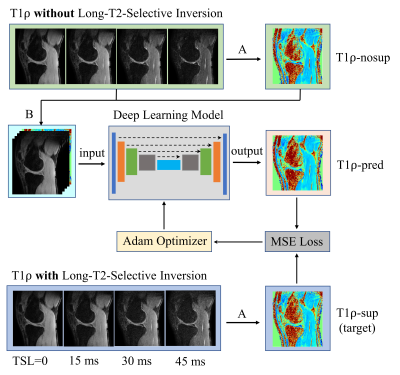
Figure 2.
Workflow for using deep learning to obtain
synovial fluid suppressed T1ρ maps from MRI scans without long-T2-selective
inversion (LT2SI). Operation A: calculation of T1ρ maps using conventional
non-linear exponential curve fitting; Operation B: stack of the T1ρ-nosup image
and the TSL source images to form a five-channel dataset as input to the deep
learning model.
