Ralf Vogel1,2, Dieter Ritter2, Jonathan Weine2,3, Jonas Faust2,4, Elodie Breton5, Julien Garnon5,6, Afshin Gangi5,6, Andreas Maier1, and Florian Maier2
1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Siemens Healthcare, Erlangen, Germany, 3TU Dortmund, Dortmund, Germany, 4Universität Heidelberg, Heidelberg, Germany, 5ICube UMR7357, University of Strasbourg, CNRS, FMTS, Strasbourg, France, 6Imagerie Interventionnelle, Hôpitaux Universitaires de Strasbourg, Strasbourg, France
1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Siemens Healthcare, Erlangen, Germany, 3TU Dortmund, Dortmund, Germany, 4Universität Heidelberg, Heidelberg, Germany, 5ICube UMR7357, University of Strasbourg, CNRS, FMTS, Strasbourg, France, 6Imagerie Interventionnelle, Hôpitaux Universitaires de Strasbourg, Strasbourg, France
Synthetic training data was successfully used to train a U-Net for image-based needle tracking, utilizing automatically generated ground-truth labels. The evaluation on clinical patient data indicates that synthetic MR images can replace patient data and animal experiments.
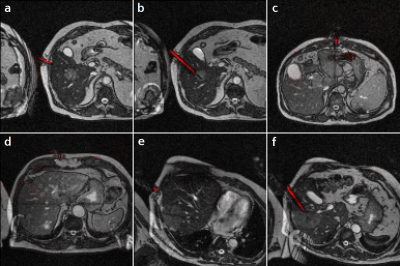
Figure
1: Needle
predictions on clinical interventional images (red overlays), generated by U-net
trained with synthetic data only.
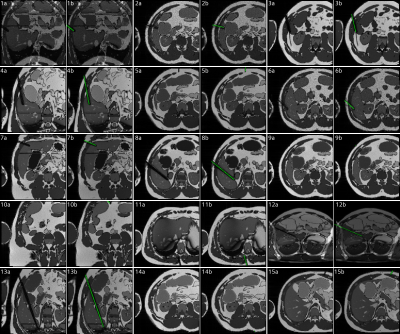
Figure
5: (a) random
selection of simulated images generated using the AustinMan model. All images
were simulated in the transversal plane, tilted by up to ±45° to the coronal plane. The needle was either placed
in liver tissue from a reasonable angle or completely random (11, 12, 14, 15). (b) Image
including the automatically generated needle label (green).
