Pan Su1,2, Sijia Guo2,3, Steven Roys2,3, Florian Maier4, Thomas Benkert4, Himanshu Bhat1, Elias R. Melhem2, Dheeraj Gandhi2, Rao P. Gullapalli2,3, and Jiachen Zhuo2,3
1Siemens Medical Solutions USA, Inc., Malvern, PA, United States, 2Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland, School of Medicine, Baltimore, MD, United States, 3Center for Metabolic Imaging and Therapeutics (CMIT), University of Maryland Medical Center, Baltimore, MD, United States, 4Siemens Healthcare GmbH, Erlangen, Germany
1Siemens Medical Solutions USA, Inc., Malvern, PA, United States, 2Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland, School of Medicine, Baltimore, MD, United States, 3Center for Metabolic Imaging and Therapeutics (CMIT), University of Maryland Medical Center, Baltimore, MD, United States, 4Siemens Healthcare GmbH, Erlangen, Germany
Deep
learning can be used to generate synthetic CT skull, thereby simplifying
workflow of tcMRgFUS. 3D V-Net can take advantage of contextual information in
volumetric images. Furthermore, the pre-trained model can be applied in dataset
acquired using different sequence/protocols.
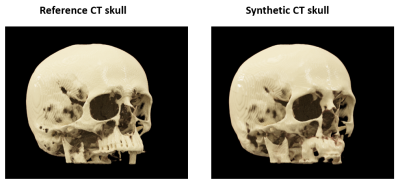
Figure
4: 3D cinematic rendering of the reference CT skull and synthetic CT skull from
a representative subject.
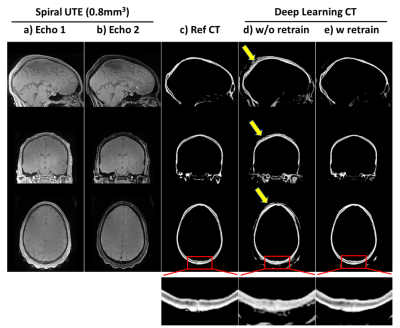
Figure 5: Transfer learning for 0.8mm3
isotropic synthetic CT skull: results of applying the trained model above to dual echo spiral UTE data (0.8mm3
isotropic) acquired on different 3T scanner. ab) dual echo spiral UTE; c) reference
CT skull; d) results of directly applying the previous model trained on radial UTE without any retraining of the spiral
UTE data; e)
results of applying the model after retraining with single spiral UTE data. Bottom
are zoom-in windows showing the details in posterior skulls. Note that all CTs here
are 0.8mm3 isotropic, as CT were coregistered to UTE space.