Digital Poster
Software Tools
ISMRM & ISMRT Annual Meeting & Exhibition • 10-15 May 2025 • Honolulu, Hawai'i

 |
Computer Number: 1
1958. Design
and evaluation of an educational MRI simulator (eduMRIsim)
S. Gonzalez Riedel, A. Raaijmakers, M. Breeuwer
Eindhoven University of Technology, Eindhoven, Netherlands
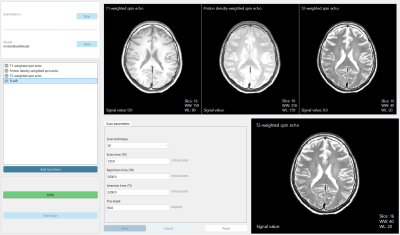
Impact: The new open-source MRI simulator, eduMRIsim,
mimics a clinical scanner allowing students to
experimentally explore scan parameter effects on image
appearance. Positive feedback from educators affirms its
potential as a valuable education tool for the next
generation of MRI researchers.
|
|
 |
Computer Number: 2
1959. Accelerated
GVE File Parsing for Philips MRI Sequences Using Julia
J. Kloiber, A. Jaffray, C. Graf, A. Rauscher
University of British Columbia, Vancouver, Canada
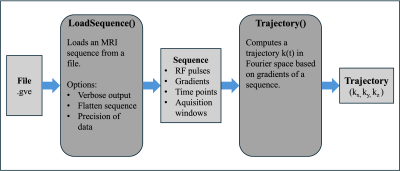
Impact: Long load times of MRI sequences can
significantly slow down simulation and reconstruction
projects. The proposed Julia module provides a fast and
accurate solution that can easily be integrated into other
projects.
|
|
 |
Computer Number: 3
1960. Software
package distribution of slicewise motion correction tool
(SLOMOCO) for fMRI pre-processing
W. Shin, P. Taylor, R. Reynolds, M. Lowe
Cleveland Clinic, Cleveland, United States
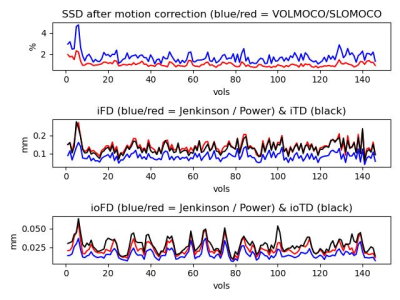
Impact: The SLOMOCO software is publicly available. We
demonstrate its usage, and the performance is validated with
10 different inter-/intra-volume motion corrupted MR
datasets using ex-vivo brain.
|
|
 |
Computer Number: 4
1961. Modular
and Open-Source Interactive Real-Time MRI
P. Schaten, N. Scholand, D. Mackner, M. Blumenthal, M.
Uecker
Graz University of Technology, Graz, Austria
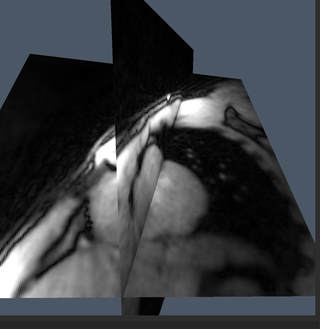
Impact: We present a comprehensive interactive real-time
MRI framework built from modular, open-source components and
geared towards interventional MRI to facilitate the use of
interactive real-time MRI in clinical applications.
|
|
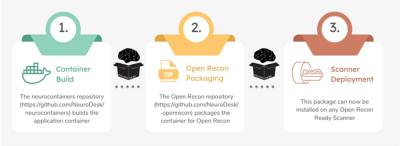 |
Computer Number: 5
1962. Enabling
Neurodesk workflows on the scanner console
M. Xu, S. Bollmann, S. Stefan, D. Nanz, P. Pullens, K. Pine,
K. Chow, R. Schneider, T. Wuerfl, M. Barth, D. Guellmar
The University of Queensland, Brisbane, Australia
Impact: By enabling seamless integration of Neurodesk
containers directly on the MRI scanner console, this work
simplifies imaging pipeline deployment, reduces data
analysis delays, and allows rapid image analysis ultimately
supporting broader accessibility and clinical translation of
advanced image processing pipelines.
|
|
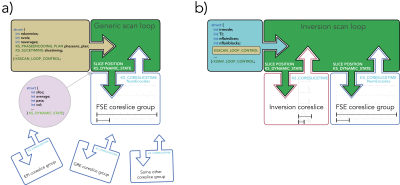 |
Computer Number: 6
1963. Generic
looping in KSFoundation: a programming abstraction for rapid
prototyping of pulse sequences
E. Avventi, H. Ryden, A. Van Niekerk, O. Norbeck, S.
Schauman, S. Skare
Karolinska university hospital, Stockholm, Sweden
Impact: The abstractions presented in this study has
been incorporated in the latest version of KSFoundation that
was recently released and is available on request [3].
|
|
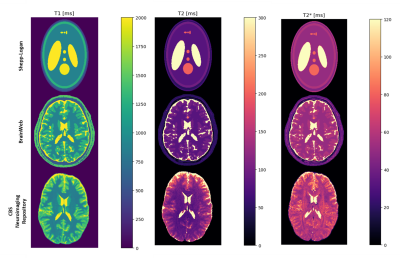 |
Computer Number: 7
1964. MRtwin:
an open-source Python package to create virtual qMRI phantoms
for benchmark and synthetic data generation
M. Cencini, M. Lancione, L. Biagi, A. Retico, M. Tosetti
INFN, Pisa Division, Pisa, Italy
Impact: MRTwin represents a useful tool for sequence
design, reconstruction optimization and benchmarking, by
providing a framework for the generation of digital twins
for quantitative imaging.
|
|
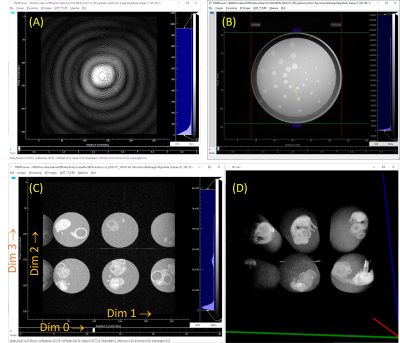 |
Computer Number: 8
1965. Python-Based
Console for Research/Metrology MRI Scanners
S. Russek, K. Stupic, K. Keenan, S. Ogier, E. Buchanan, E.
Emery
NIST, Boulder, United States
Impact: The open Python console wrapper will allow
easier development of new pulse sequences, assessment of
MRI-measurement biases, better integration of the scanner
with other required hardware, and provide a simple path to
Python-based image processing.
|
|
 |
Computer Number: 9
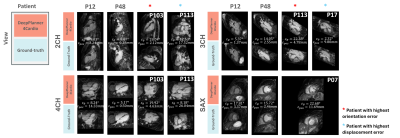
1966. DeepPlanner4Cardio:
an automatic multi-view planning tool for CMR
J. Santos, P. Osório, M. Henningsson, R. Nunes, T. Correia
Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
Impact: DeepPlanner4Cardio effectively supports CMR
operators by providing an accessible, automated solution to
enable fast and reproducible cardiac view planning,
demonstrating great potential to be applied in a clinical
setting.
|
|
 |
Computer Number: 10
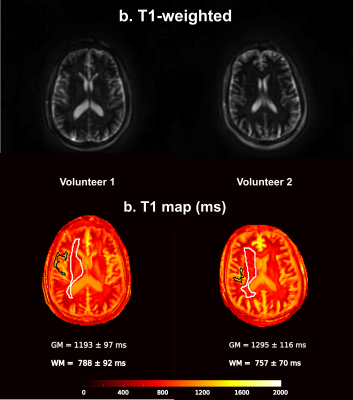
1967. In
vivo T1-mapping using Open-MOLLI in GE scanners
A. Gaspar, P. Hughes, J-F Nielsen, N. Stewart, J. Wild, T.
Correia, R. Nunes
Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
Impact: The open-source myocardial T1 mapping
(Open-MOLLI) was successfully adapted and tested on a GE
Signa 3T PET-MR scanner. The sequence is now ready to be
applied in inter-scanner reproducibility brain studies.
Triggering will be added next for cardiac applications.
|
|
 |
Computer Number: 11
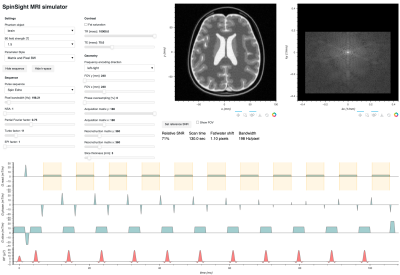
1968. SpinSight
– An educational open-source MRI simulator with joint
visualization of pulse sequence, k-space, and MR image
J. Berglund, K. Jain, J. Sousa, K. Hedman, M. Fahlström
Medical Physics, Uppsala University Hospital, Uppsala, Sweden
Impact: An open-source browser-based educational MRI
simulator was developed that jointly visualizes four levels
of the imaging process: acquisition parameters, pulse
sequence, k-space, and the MR image. Low latency enables
students to explore the interaction between these levels in
real time.
|
|
 |
Computer Number: 12
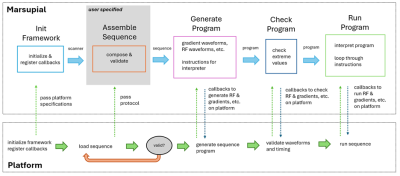
1969. MR
Sequence Utility for Portable Imaging and Abstraction Layers
(Marsupial) – an MR Sequence Programming Framework written in
Nim
A. Petrovic, R. Bammer
Monash Health, Melbourne, Australia
Impact: Marsupial will enable development of
platform-independent open-source MRI sequences. Nim’s
concise syntax and Marsupial’s design will considerably
speed up the development process, making more sequences
readily available to the research community.
|
|
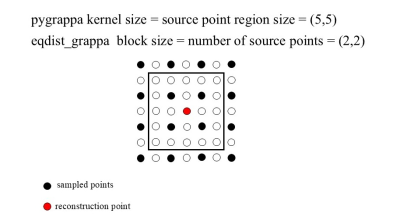 |
Computer Number: 13
1970. A
fast Pytorch based GRAPPA implementation for uniformly
undersampled k-space
Y. Bu, Z. Wang, J. Li, Y. Liu, M. Lyu
Shenzhen Technology University, Shenzhen, China
Impact: This approach enables faster MRI
reconstructions, making it suitable for many applications.
|
|
 |
Computer Number: 14
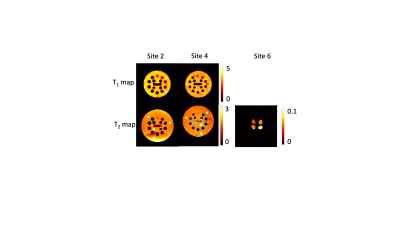
1971. Multi-site
use of a vendor-agnostic, open-source protocol (VOP) framework
to validate and share open-source pulse sequences
S. Geethanath, A. Artiges, T. Block, Q. Chen, T. Fernandes,
S. Ganji, W. Grissom, D. Hoinkiss, J. Vaughan Jr., A. Konar,
S. Konstandin, V. Mascarenhas, M. Nagtegaal, J. Nielsen, R.
Nunes, M. Shafiekhani, M. Zaitsev
Johns Hopkins University, Ellicott City, United States
Impact: We demonstrated the expanded use of the VOP
framework to validate and share the IRSE and TSE sequences
across sites, vendors, software versions, and two field
strengths. This framework created an online platform to
interact and share pulse sequences.
|
|
 |
Computer Number: 15
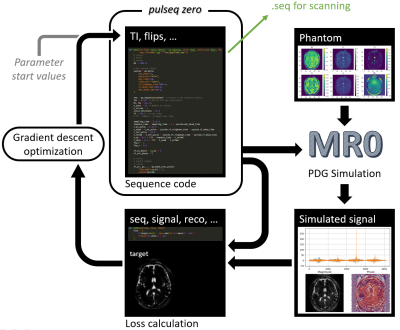
1972. Pulseq-zero:
PyPulseq sequence scripts in a differentiable optimization loop
J. Endres, M. Zaiss
University Clinic Erlangen, Erlangen, Germany
Impact: Pulseq is a widely used and vendor agnostic
library for writing MRI sequences. With pulseq-zero,
existing and new sequence scripts can be simulated and
optimized directly, with only minor modifications. The
resulting sequences can be measured on real scanners
immediately.
|
|
 |
Computer Number: 16
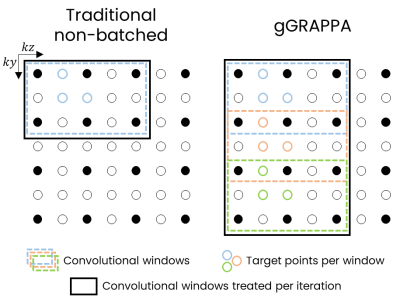
1973. gGRAPPA:
A Flexible, GPU-Accelerated Python Package for Fast and
Efficient Generalized GRAPPA Reconstruction
M. Bertrait, C. R, P. Ciuciu
CEA Neurospin, Gif-sur-Yvette, France
Impact: gGRAPPA provides a fast, flexible, and
open-source solution for GRAPPA MRI reconstruction on GPU,
significantly accelerating reconstruction times and enabling
ultra high-resolution imaging reconstruction, thereby
supporting advanced research applications across diverse MRI
protocols.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.