Digital Poster
AI for Image Segmentation: Above the Neck
ISMRM & ISMRT Annual Meeting & Exhibition • 10-15 May 2025 • Honolulu, Hawai'i

 |
Computer Number: 33
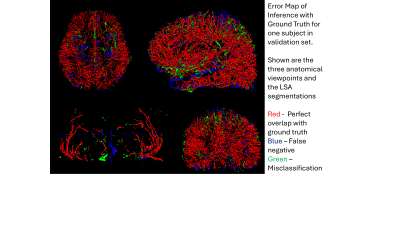
1849. Deep
learning segmentation of small blood vessels and vessel density
mapping based on high resolution black blood MRI
S. Mendoza, Z. Yang, J. Lamas, K. Jann, M. Harrington, J.
Ringman, Y. Shi, D. Wang
USC, Los Angeles, United States
Impact: A robust cerebral small vessel density
estimation method can help for large-scale analysis of black
blood MRI data to look for possible biomarkers of early
cognitive decline. Our pipeline will be shared with the
community.
|
|
 |
Computer Number: 34
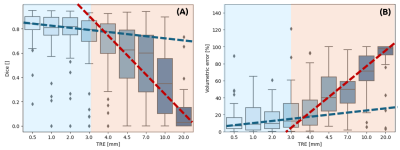
1850. Study
of the impact of registration errors on the segmentation of
stroke lesion in deep learning.
O. Pulvéric, F. Ouadahi, T. Boutelier
Olea Medical, La Ciotat, France
Impact: By identifying a critical 3 mm threshold for
registration errors, we established quantitative guidelines
for acceptable registration accuracy in clinical workflows.
This threshold helps define minimal performance requirements
for registration algorithms when preprocessing images for
segmentation tasks.
|
|
 |
Computer Number: 35
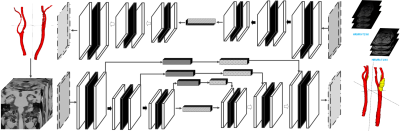
1851. Deep
Learning-Driven Architecture for Automated Segmentation and
High-Risk Carotid Plaque Identification in High-Resolution MRI
X. Cao, Z. Zheng, Q. Yang
Academy for Engineering and Technology, Fudan University, Shanghai, China
Impact: This study significantly enhances the
diagnostics of carotid atherosclerosis by leveraging
high-resolution MRI and deep learning to accurately identify
high-risk plaques, improving early intervention and
potentially transforming outcomes in cardiovascular patient
care.
|
|
 |
Computer Number: 36
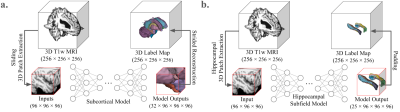
1852. Improving
Subcortical and Hippocampal Subfield Segmentation with a 3D
Hybrid Deep Learning Solution
A. Cao, Z. Li, J. Jomsky, A. Laine, J. Guo
University of California, Santa Barbara, San Jose, United States
Impact: Our proposed novel deep learning model,
MedSegMamba, reliably demonstrated state-of-the-art
segmentation performance and utility across numerous
datasets. It outperformed other well-established deep
learning tools on the difficult tasks of subcortical and
hippocampal subfield segmentation. Code is available
here: https://github.com/aaroncao06/MedSegMamba.
|
|
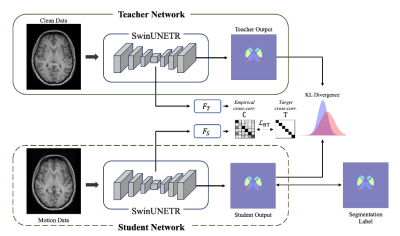 |
Computer Number: 37
1853. Improving
Subcortical Segmentation in Brain MRI Using Knowledge
Distillation to Enhance Robustness Against Motion Artifacts
C. Ryu, S. Jung, Y. Choi, D-H Kim
Yonsei University, Seoul, Korea, Republic of
Impact: This approach improves MRI segmentation of
motion-corrupted data, supporting reliable subcortical
analysis without complex preprocessing. It provides a method
for cleaner, artifact-resistant segmentation, presenting
significant applications both in neurodevelopmental and
neurodegenerative disease research.
|
|
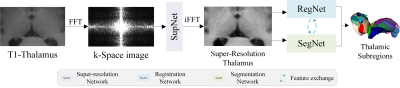 |
Computer Number: 38
1854. A
k-Space Super-Resolution and Registration-Guided Thalamic
Subregions Segmentation Model for Analyzing Thalamic Iron
Changes in AD
J. He, D. Li, B. Fu, L. Nie, R. Wang
Guizhou Provincial People’s Hospital, Guiyang, China
Impact: There is currently a lack of research on
quantitative iron analysis in fine-grained thalamic
subregions of AD patients. This study could provide new
prognostic assessments and therapeutic target references for
AD research.
|
|
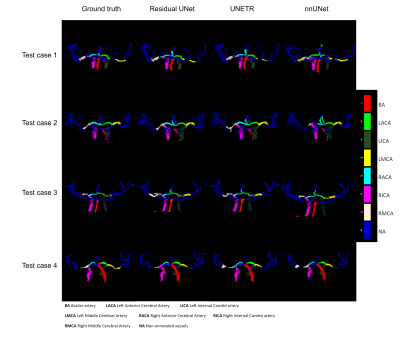 |
Computer Number: 39
1855. Automatic
Labelling of Intracranial arteries: A Comparison of UNet-based
Networks
J. Bisbal, S. Jofré, P. Winter, A. Ponce, M. Aristova, J.
Moore, O. Welin Odeback, S. Ansari, C. Tejos, S. Uribe, M.
Markl, J. Sotelo, S. Schnell, D. Marlevi
Pontificia Universidad Catolica de Chile, Santiago, Chile
Impact: This study identifies Residual UNet as an
effective tool for automated intracranial artery labeling,
enabling fully automatic processing of intracranial 4D Flow
MRI data.
|
|
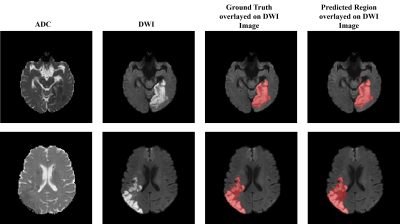 |
Computer Number: 40
1856. A
Two Step Deep Learning Framework for Identifying Ischemic Stroke
Core: Integration of Inception-v3 and MultiResU-Net on DWI and
ADC MRI Images
A. Kandpal, P. Prajapati, S. Maurya, R. Gupta, A. Singh
Indian Institute of Technology Delhi, New Delhi, India
Impact: The proposed deep-learning framework combines
DWI images and ADC maps to enable automatic, rapid ischemic
core segmentation with high accuracy, supporting
radiologists in the objective evaluation and planning of
stroke treatment.
|
|
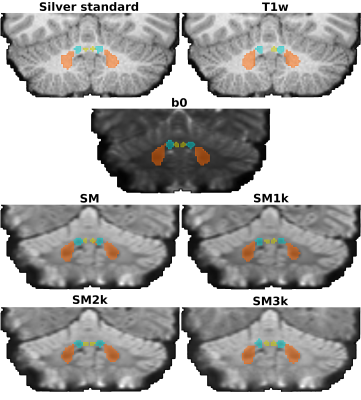 |
Computer Number: 41
1857. Diffusion
MRI spherical mean improves deep cerebellar nuclei segmentation
J. H. Legarreta, Z. Lan, Y. Chen, F. Zhang, E. Yeterian, N.
Makris, J. Rushmore, Y. Rathi, L. O’Donnell
Brigham and Women's Hospital, Mass General Brigham/Harvard Medical School, Boston, United States
Impact: Diffusion MRI spherical mean should be
considered as a relevant image contrast for cerebellar
structure segmentation using deep neural networks towards an
increasingly accurate connectivity analysis.
|
|
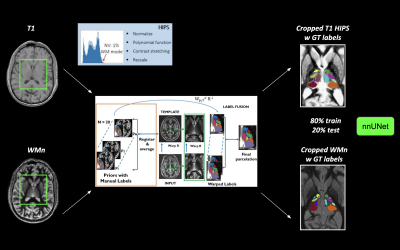 |
Computer Number: 42
1858. Deep-THOMAS:
a robust deep learning network for fast and accurate thalamic
nuclei segmentation
A. Barat, R. Ramesh, A. Cacciola, A. Banerjee, M. Saranathan
University of Massachusetts Amherst, Amherst, United States
Impact: This robust thalamic nuclei segmentation tool
can be integral for clinical neuroimaging, offering fast,
reliable segmentation, opening the door for analysis of
large public databases to study the role of thalamic nuclei
in a variety of neurodegenerative and neuropsychiatric
conditions.
|
|
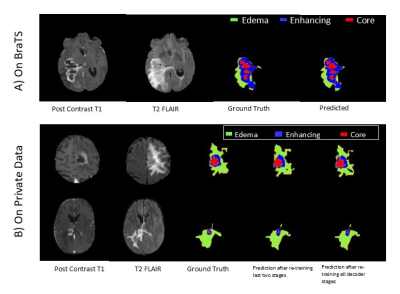 |
Computer Number: 43
1859. Comparative
Analysis of Deep Learning Models for Brain Tumor Segmentation in
MRI Scans Using BraTS and Experimental Datasets
S. Misra, S. Bera, S. Basak, S. Sarkar, A. Rajan, S. Mohan,
H. Poptani, S. Chawla, S. Bhaduri
TCG Centres for Research & Education in Science & Technology, Kolkata, India
Impact: It highlights potential of deep-learning models,
particularly nnU-Net to improve accuracy and efficiency in
brain-tumor segmentation, reducing reliance on
labor-intensive methods like RANO and iRANO. Addressing
dataset-specific limitations through transfer-learning, the
findings aids in consistent tumor-volume assessment,
enhancing treatment monitoring.
|
|
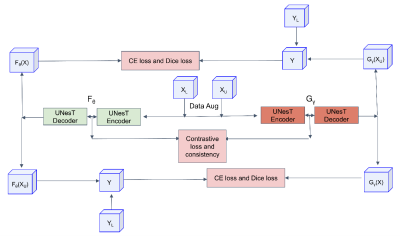 |
Computer Number: 44
1860. Contrastive
Mutual Learning: A Semi-Supervised Method for 3D Fetal Brain
Segmentation
S. Li, L. Jia, W. Cai, C. Wang, H. Wang, H. Li
Institute of Science and Technology for Brain-inspired Intelligence, Fudan University, Shanghai, China, Shanghai, China
Impact: This study implemented a semi-supervised
learning approach to address the challenges of limited
labeled data and high annotation costs in 3D fetal brain
segmentation. The performance demonstrates the proposed
algorithm's robust potential.
|
|
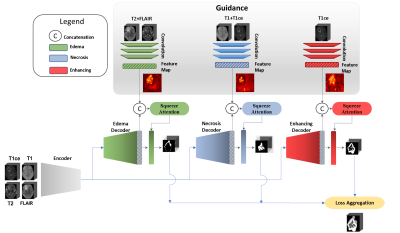 |
Computer Number: 45
1861. Improving
Brain Tumor Segmentation with a Clinically-Informed
Multi-Decoder U-Net
A. Rezk, A. Al-Fakih, A. Shazly, K. Ryu, M. A. Al-masni
Sejong University, Seoul, Korea, Republic of
Impact: Our clinically guided, multi-decoder U-Net
demonstrates improved segmentation accuracy, particularly in
diverse and non-standard datasets. This innovation paves the
way for more reliable, adaptable, and interpretable brain
tumor imaging, enhancing diagnostic confidence and treatment
planning.
|
|
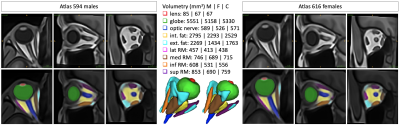 |
Computer Number: 46
1862. MR-Eye
atlas: a large-scale atlas of the eye based on T1-weighted MR
imaging
J. Barranco, A. Luyken, P. Stachs, O. Esteban, Y. Aleman, S.
Langner, O. Stachs, B. Franceschiello, M. Bach Cuadra
CIBM Center for Biomedical Imaging, Lausanne, Switzerland
Impact: The publicly provided large-scale unbiased T1w
MR-Eye atlases will facilitate spatial normalization and
quantitative analysis in the field of ophthalmic imaging,
helping clinicians in the diagnosis of multiple ocular
diseases, and enhancing our understanding in sex-specific
eye anatomy and physiology.
|
|
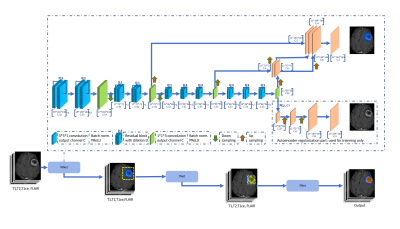 |
Computer Number: 47
1863. Automated
Pipeline Development for Multi-Compartmental Volumetric
Glioblastoma Segmentation and Advanced MRI Parameter Extraction
E. Lotan, A. Saulnier, J. Nguyen, E. Hammon, A. Davis, M.
Lee
NYU Grossman School of Medicine, New York, United States
Impact: This automated pipeline can enhance clinical
decision-making and personalized treatment for glioblastoma
patients. Its development will facilitate new research on
imaging biomarkers, ultimately improving patient outcomes
and advancing neuroimaging practices.
|
|
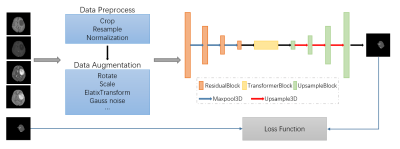 |
Computer Number: 48
1864. A
Comprehensive Deep Learning Approach for Multi-type Central
Nervous System Tumor Segmentation Based on the 2021 WHO
Classification
S. Li, L. Guo, Y. Tian, G. Ju, S. Zhang, Y. Jin, X. Su, S.
Tang, A. Zeng, Y. Luo, X. Yang, L. Wang, L. Wang, H. Zhang,
W. Yang, X. Liang, Q. Yue
West China Hospital of Sichuan University, Chengdu, China
Impact: This work advances automated, multi-tumor
segmentation tools, enhancing clinical workflows and
supporting consistent, efficient CNS tumor analysis.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.