Digital Poster
Processing & Analysis: Segmentation II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
2223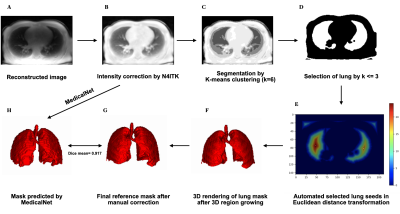 |
36 | Lung segmentation with deep learning for 3D MR spirometry
Zhongzheng He1,2, Nathalie Barrau1, Claire Pellot Barakat1, and Xavier Maître1
1Université Paris-Saclay, CEA, CNRS, Inserm, BioMaps, Orsay, France, 2IADI U1254, INSERM, Université de Lorraine, Nancy, France
Current MR lung image segmentation has huge challenges compared to CT images, specifically in terms of low contrast, non-homogeneity. We developed a series of processing to accelerate the manual correction of reference standard lung masks by the auto-seeds region growing. Finally, we developed a 3D automatic MRI lung segmentation method using deep learning with a limited dataset (41 volumes for training and 4 volumes for validation). In the primary result, this lung segmentation archived a Dice score of 0.917±0.013. In the case of limited data, it provides us a new way for MR lung segmentation.
|
||
2224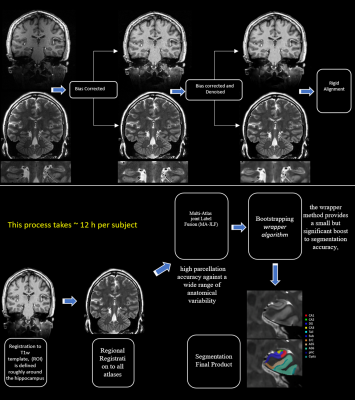 |
37 | Hippocampal Subfields Volume in Middle Age Healthy Adults Video Not Available
salem Alkhateeb1, Tales Santini1, Li jinghang1, Robin Chu1, Daniel Ibrahim1, Anna M. Marsland1, Stephen B. Manuck1, Pete Gianaros1, and tamer S. Ibrahim2
1University of Pittsburgh, Pittsburgh, PA, United States, 2University of Pittsburgh, pittsburgh, PA, United States
As hippocampal volume has been extensively utilized as a diagnosing tool to confirm diagnosis of many neurological disorders, this study aims to employ the high resolution 7T TSE T2w data to segment high quality images with precision based on multi atlases and machine learning. Results have proved that this pipeline provides excellent outcomes and is validated to be used for more variables.
|
||
2225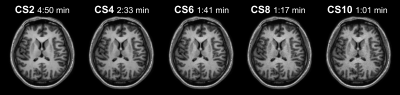 |
38 | Effect of Compressed SENSE on Freesurfer parcellation precision
Michael A Green1,2, Peter Humberg3, Iain K Ball4, and Caroline D Rae1,2
1Neuroscience Research Australia, Sydney, Australia, 2School of Medical Sciences, University of New South Wales, Sydney, Australia, 3Stats Central, Mark Wainwright Analytical Centre, University of New South Wales, Sydney, Australia, 4Philips Australia & New Zealand, Sydney, Australia
We employed the commonly used software package, Freesurfer to obtain volume, area and thickness measurements from structural MR imaging data acquired using the Compressed SENSE acceleration technique. We assessed measurement reliability via equivalence testing on a series of increasing acceleration factors to provide guidelines for researchers wanting to reduce scan acquisition time while acquiring high-quality data.
|
||
2226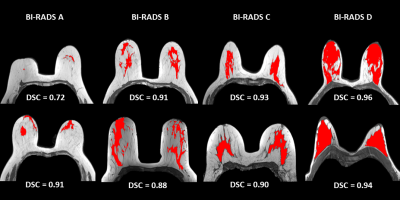 |
39 | Automatic fibroglandular tissue segmentation in breast MRI using a deep learning approach
Fares Ouadahi1, Anais Bernard1, Lucile Brun1, and Julien Rouyer1
1Department of Research & Innovation, Olea Medical, La Ciotat, France
An accurate fibroglandular tissue (FGT) segmentation model was designed using of a deep learning strategy on T1w series without fat suppression. The proposed method combined a dedicated preprocessing and the training of a two-dimensional U-Net architecture on a multi-centric representative database to achieve an automatic FGT segmentation. The final test of the generated model exhibited overall good performances with a median Dice similarity coefficient of 0.951. More contrasted performances were obtained when correlating the gland density with the discrepancy between ground truth and prediction. Indeed, the lower the breast density, the greater the uncertainty in the segmentation.
|
||
2227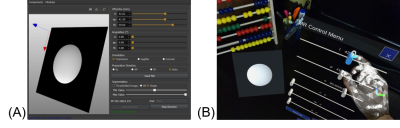 |
40 | Immersive and Interactive On-the-fly MRI Control and Visualization with a Holographic Augmented Reality Interface: A Trans-Atlantic Test
Jose Velazco-Garcia1, Nikolaos Tsekos1, Andrew Webb2, and Kirsten Koolstra2
1Medical Robotics and Imaging Lab, University of Houston, Houston, TX, United States, 2Leiden University Medical Center, Leiden, Netherlands
We present the computational Framework for Interactive Immersion into Imaging Data (FI3D) for 3D visualization of MR data as they are collected and on-the-fly control of the MRI scanner. The FI3D was implemented to integrate an MRI scanner with the operator who used a customized holographic scene, generated by a HoloLens head mounted display (HMD), to (1) view images and image-generated outcomes (e.g., segmentation) and (2) control the MR scanner to update the acquisition protocol. In this study, the framework linked an MR scanner in Leiden (The Netherlands) and an operator in Houston (USA).
|
||
2228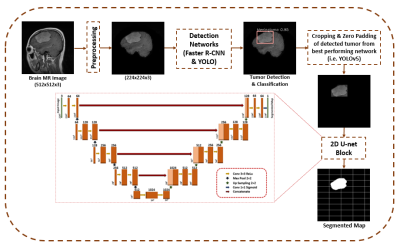 |
41 | A Framework for Brain Tumor Detection, Classification and Segmentation using Deep Learning
Rafia Ahsan1, Iram Shahzadi2,3, Ibtisam Aslam1,4, and Hammad Omer1
1Medical Image Processing Research Group (MIPRG), Dept. of Elect. & Comp. Engineering, COMSATS University Islamabad, Islamabad, Pakistan, 2OncoRay – National Center for Radiation Research in Oncology, Faculty of Medicine and University Hospital Carl Gustav Carus, Technische Universität Dresden, Helmholtz-Zentrum Dresden – Rossendorf, Dresden, Germany, 3German Cancer Research Center (DKFZ), Heidelberg, Germany, 4Service of Radiology, Geneva University Hospitals and Faculty of Medicine, University of Geneva, Geneva, Switzerland
Detection, classification and segmentation of brain tumor simultaneously is challenging due to the heterogeneous nature of the tumor. Limited work has been done in literature in this regard. The present study, therefore, aims to identify an object detection network that would be able to solve multi-class brain tumor classification and detection problem with high accuracy. Furthermore, the best performing detection network has been cascaded with 2D U-Net for pixel level segmentation. The proposed method not only classifies the tumor with high accuracy but also provides improved segmentation results compared to the standard U-Net.
|
||
2229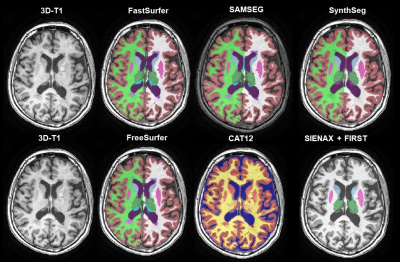 |
42 | Cross-sectional robustness of 6 freely available software packages for brain volume measurements in multiple sclerosis Video Permission Withheld
David Rudolf van Nederpelt1, Houshang Amiri1,2, Amanda Mariyampillai1, Iman Brouwer1, Samantha Noteboom3, Frederik Barkhof1,4, Joost P.A. Kuijer1, and Hugo Vrenken1
1Radiology and nuclear medicine, Amsterdam UMC, location VUmc, Amsterdam, Netherlands, 2Neuroscience research center, Kerman University of Medical Sciences, Kerman, Iran (Islamic Republic of), 3Anatomy and Neurosciences, Amsterdam UMC, location VUmc, Amsterdam, Netherlands, 4Institutes of Neurology and Healthcare Engineering, UCL London, London, United Kingdom
Automated segmentation of brain MR images has paved the way for large cohort atrophy studies in multiple sclerosis (MS). A variety of automated software packages is available. Here we aimed to quantify brain volume differences measured by six freely available software packages on data from 21 MS patients all scanned thrice on three different MR scanners. While intra-class correlation coefficients were high, systematic differences between scanners were found for every software. This suggests that direct comparison of volumes acquired with different scanners is not possible and standardization is needed.
|
||
2230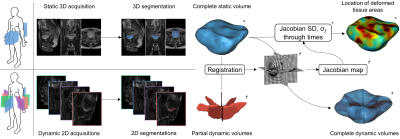 |
43 | Three-dimensional reconstruction and characterization of bladder deformations
Augustin C. Ogier1, Stanislas Rapacchi2, and Marc-Emmanuel Bellemare1
1Aix Marseille Univ, Universite de Toulon, CNRS, LIS, Marseille, France, 2Aix Marseille Univ, CNRS, CRMBM, Marseille, France Pelvic floor disorders are prevalent diseases and only 2D dynamic observations of straining exercises at excretion are available in clinics. The understanding of three-dimensional pelvic organs mechanical defects is not yet achievable. We proposed a complete methodology for the 3D representation of the bladder combined with 3D representation of the location of the highest strain areas on the organ surface. We assessed the potential of our method on eight control subjects for the reconstruction of bladder during intense straining from forced breathing exercises. The proposed methodology provided, for the first time, a proper 3D+t spatial tracking of bladder non-reversible deformations. |
||
2231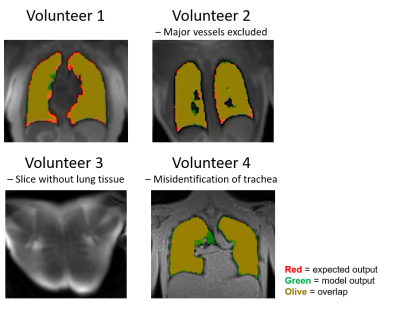 |
44 | Deep Learning-Based Automatic Lung Segmentation for MR Images at 0.55T
Rachel Chae1,2, Ahsan Javed1, Rajiv Ramasawmy1, Hui Xue1, Marcus Carlsson1, Adrienne E Campbell-Washburn1, and Felicia Seemann1
1Cardiovascular Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, United States, 2Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States
High-performance 0.55T systems are well suited for structural lung imaging due to reduced susceptibility and prolonged T2*. Lung segmentation is required to derive metrics of pulmonary function from lung MRI. Convolutional neural networks are effective for lung segmentation at higher field strengths, but they do not generalize to images acquired at 0.55T. This study develops a neural network for automated lung segmentation of T1 and proton density weighted ultrashort-TE MRI at 0.55T. Training data was generated using segmentations by active contours and manual corrections. The proposed network was fast (1.07s) and as accurate as existing semi-automated segmentation (dice coefficient 0.93).
|
||
2232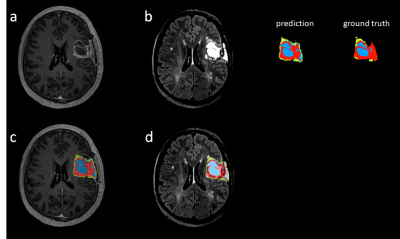 |
45 | Deep learning-based tumor segmentation from postoperative MRI
Jingpeng Li1,2, Jonas Vardal3,4, Inge Groote1, and Atle Bjornerud1,2
1Computational Radiology and Artificial Intelligence, Oslo University Hospital, Oslo, Norway, 2Department of Physics, University of Oslo, Oslo, Norway, 3The Intervention Centre, Oslo University Hospital, Oslo, Norway, 4Faculty of Medicine, University of Oslo, Oslo, Norway
To accurately detect and localize postoperative tumor after surgery is of critical importance to postoperative patient management and survial rate. We propose a fully automated end-to-end coarse-to-fine segmentation approach for the segmentation of posoperative tumor.
|
||
2233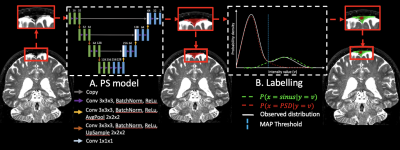 |
46 | Deep learning-based segmentation of the parasagittal dural space from non-contrast anatomical MRI image: a resource for glymphatic studies
Kilian Hett1, Colin D. McKnight2, Jarrod J. Eisma1, Jason Elenberger1, Ciaran M. Considine1, Daniel O. Claassen1, and Manus J. Donahue1,2,3
1Neurology, Vanderbilt University Medical Center, Nashville, TN, United States, 2Radiology and Radiological Sciences, Vanderbilt University Medical Center, Nashville, TN, United States, 3Psychiatry and Behavioral Sciences, Vanderbilt University Medical Center, Nashville, TN, United States The overarching goal of this work is to develop and validate novel deep learning algorithms for segmenting the parasagittal dural (PSD) space, which has been hypothesized to harbor cerebral lymphatic channels, from standard non-contrast anatomical imaging. Specifically, contrasted based MRI studies have recently suggested that the PSD may be important for CSF clearance, however existing methods for evaluating this space require administration of exogenous contrast and time-consuming manual tracing, thereby limiting generalizability. We propose a new segmentation method using non-contrasted MRI, and we validate this method in a mixed cohort of older adults with and without neurodegeneration. |
||
2234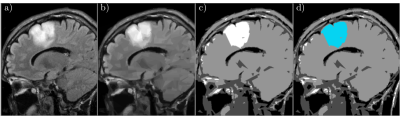 |
47 | Split-and-Merge Segmentation in Magnetic Resonance Imaging Based on Graph Wedgelets Video Permission Withheld
Wolfgang Erb1
1Dipartimento di Matematica "Tullio Levi-Civita", University of Padova, Padova, Italy
Graph wedgelets are a novel tool for the fast decomposition of images in geometrically meaningful, wedge-shaped subregions. In this work, we study the usage of graph wedgelets as a promising splitting method in a split-and-merge segmentation scheme for Magnetic Resonance Imaging. We combine adaptive wedgelet splits of MRI images with a simple and classical merging strategy for subregions and obtain in this way an efficient and robust segmentation of diagnostic-relevant subdomains in MRI data.
|
||
2235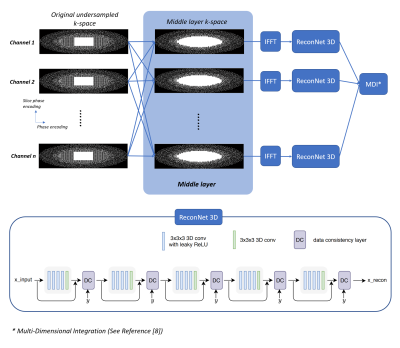 |
48 | Cascaded parallel imaging for 3D deep learning reconstruction
Jingyuan Lyu1, Yongquan Ye1, Zhongqi Zhang1, Jian Xu1, Xiao Chen2, Terrence Chen2, Shanhui Sun2, and Eric Z. Chen2
1UIH America, Inc., Houston, TX, United States, 2United Imaging Intelligence, Cambridge, MA, United States
This study demonstrates the effects on image reconstruction by integrating a separate parallel imaging layer with the deep learning module. With such additional layer, deep learning reconstruction is flexible and reliable for highly under-sampled data.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.