Digital Poster
Processing & Analysis: Software Tools & Visualization I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
2682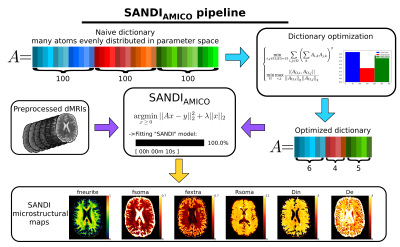 |
46 | SANDIAMICO: an open-source toolbox for Soma And Neurite Density Imaging (SANDI) with AMICO
Simona Schiavi1,2, Mario Ocampo-Pineda1, Raphaël Truffet3, Emmanuel Caruyer3, Alessandro Daducci1, and Marco Palombo4,5,6
1Department of Computer Science, University of Verona, Verona, Italy, 2Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health (DINOGMI), University of Genoa, Genoa, Italy, 3CNRS, Inria, Inserm, Univ Rennes, Rennes, France, 4Centre for Medical Image Computing (CMIC), Department of Computer Science, University College London, London, United Kingdom, 5Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 6School of Computer Science and Informatics, Cardiff University, Cardiff, United Kingdom The soma and neurite density imaging (SANDI) model has been recently introduced to estimate diffusion MRI indices of apparent neurite and soma density noninvasively in the brain. Here, we introduce SANDIAMICO a fast (10 seconds for whole brain images) and robust implementation for estimating SANDI parameters using the Accelerated Microstructure Imaging via Convex Optimization (AMICO) framework. Using numerical simulations and in vivo human data, we show excellent performances of SANDIAMICO in terms of accuracy, precision, robustness to noise and intra-subject reproducibility. |
||
2683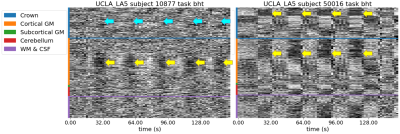 |
47 | Quality control and nuisance regression of fMRI, looking out where signal should not be found
Céline Provins1, Christopher J. Markiewicz2, Rastko Ciric2, Mathias Goncalves2, Cesar Caballero-Gaudes3, Russell A. Poldrack2, Patric Hagmann1, and Oscar Esteban1
1Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 2Department of Psychology, Stanford University, Stanford, CA, US, Stanford, CA, United States, 3Basque Center on Cognition, Brain and Language, Donostia, Spain, Donostia, Spain
Quality control of functional MRI data is essential as artifacts can have a critical impact on subsequent analysis. Yet, visual assessment of a dataset is tedious and time-consuming. By extending the carpet plot with the voxels located on a closed band (or “crown”) around the brain, we showed that fMRI data quality can be assessed more effectively. This new feature has been incorporated into MRIQC and fMRIPrep. In addition, a new nuisance regressor has been added to the latter, calculated from timeseries within this new “crown”.
|
||
2684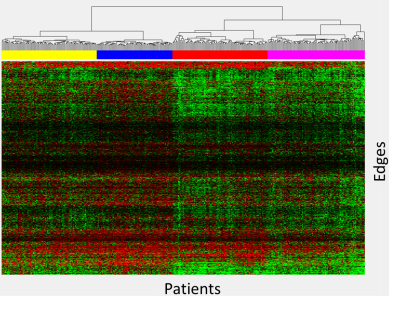 |
48 | Data-driven heatmap clustering approach to analyzing structural connectomes
Wilburn E Reddick1, Jordan Teague1, Ruitian Song1, John O Glass1, and Lisa M Jacola2
1Diagnostic Imaging, St. Jude Children's Research Hospital, Memphis, TN, United States, 2Psychology, St. Jude Children's Research Hospital, Memphis, TN, United States
Heatmap differences between 336 patients treated for ALL and 59 age-similar controls were calculated for a limited set of reproducible edges and clustered using an agglomerative hierarchical clustering that identified four clusters with unique connectivity. Differences in edges demonstrated patterns of lower (group 2), higher (group 4), or mixed (group 1 & 3) connectivity relative to controls. Neurocognitive performance was substantially below normal on measures of memory (DSF – groups 2 & 3; DSB - group 1; Visual matching – group 2). This data driven approach was able to identify four distinct groups of patients with unique connectivity profiles and cognitive performance.
|
||
2685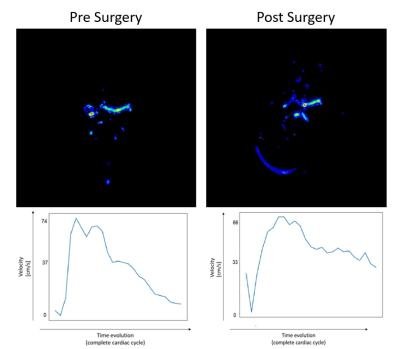 |
49 | Acquisition and finite element analysis of 4DFlow-MR functional angiography in brain and neck
María Paula Del Pópolo1,2, Leandro Enrique Salcedo1,2,3, Clara Lisazo1,2, Federico Julián González Nicolini2,4,5, Trinidad González Padin1,2, Rodrigo Nahuel Alcalá Marañón1,2,6, Rocío Paula Boschi2,7, Mercedes Paula Caspi2,7, Ezequiel F Petra8, Pedro Pablo Ariza1,2, Valdir Fialkowski9, and Daniel Fino Villamil1,2,5
1MRI Department, Fundación Argentina para el Desarrollo en Salud, Mendoza, Argentina, 2MRI Department, Fundación Escuela de Medicina Nuclear, Mendoza, Argentina, 3FI, Universidad de Mendoza, Mendoza, Argentina, 4FGQNYCS, Comisión Nacional de Energía Atómica, Buenos Aires, Argentina, 5Instituto Balseiro, Universidad Nacional de Cuyo, San Carlos de Bariloche, Argentina, 6FCEN, Universidad Nacional de Cuyo, Mendoza, Argentina, 7MRI Department, Fundación Argentina para el Desarrollo en Salud, Mendoza, Argentina, 8Hemodynamics, Fundación Argentina para el Desarrollo en Salud, Mendoza, Argentina, 9Research Department, Philips Healthcare, Barueri/San Paulo, Brazil The abscence of an angiographic functional analysis of the head and neck regions opens the possibility of applying the 4D Flow technique to quantify hemodynamics and blood flow parameters. Through a Python algorithm using Finite Elements Method the dynamic blood behavior was analyzed, obtaining a complete correlation with the clinical diagnosis. |
||
2686 |
50 | SynthStrip: skull stripping for any brain image
Andrew Hoopes1, Jocelyn S. Mora1, Adrian Dalca1,2,3, Bruce Fischl*1,2,3, and Malte Hoffmann*1,2
1Martinos Center for Biomedical Imaging, Boston, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Computer Science and Artificial Intelligence Lab, Massachusetts Institute of Technology, Cambridge, MA, United States
The removal of non-brain signal from MR data is an integral component of neuroimaging streams. However, popular skull-stripping utilities are typically tailored to isotropic T1-weighted scans and tend to fail, sometimes catastrophically, on images with other MRI contrasts or stack-of-slices acquisitions that are common in the clinic. We propose SynthStrip, a flexible tool that produces highly accurate brain masks across a landscape of neuroimaging data with widely varying contrast and resolution. We implement our method by leveraging anatomical label maps to synthesize a broad set of training images, optimizing a robust convolutional network agnostic to MRI contrast and acquisition scheme.
|
||
2687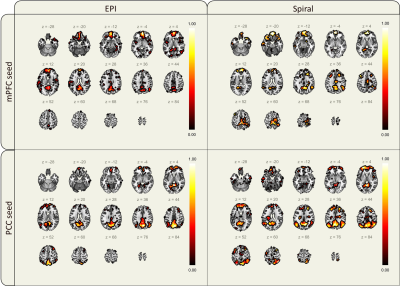 |
51 | Demonstration of an end-to-end open-source pipeline for a spiral-based rs-fMRI sequence
Marina Manso Jimeno1,2, John Thomas Vaughan Jr.1,2, and Sairam Geethanath2
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, New York, NY, United States Spiral trajectories are suitable for fMRI as they provide high temporal resolution and motion tolerance. However, they are not typically available on scanners as the associated reconstruction is more involved. We have demonstrated the feasibility of an open-source spiral sequence end-to-end pipeline and compared it to the vendor-supplied EPI sequence. Although the spiral images showed signal loss near the frontal sinus, both datasets presented similar image quality and contrast. Importantly, the spiral sequence had a 2.3-fold advantage on temporal SNR and achieved analogous functional connectivity maps of the DMN, demonstrating the sequence BOLD sensitivity and viability.
|
||
2688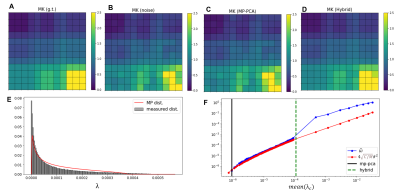 |
52 | Hybrid PCA denoising - improving PCA denoising in the presence of spatial correlations
Rafael Neto Henriques1, Sune Nørhøj Jespersen2,3, and Noam Shemesh1
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Clinical Institute, Aarhus University, Aarhus, Denmark, 3Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark
PCA denoising based on the Marchenko-Pastur (MP) distribution has become the state-of-the-art procedure to suppress thermal noise in multi-dimensional MRI. Here we developed a Hybrid-PCA strategy that combines a-priori noise variance estimation and the random matrix theory for PCA eigenvalue classification, to overcome shortcomings of contemporary MP-PCA denoising. Our results show that, while the MP-PCA denoising fails to classify the noise PCA components in data with spatially correlated noise, the Hybrid-PCA algorithm maintains its denoising performance. The Hybrid-PCA denoising can thus be a useful procedure for data corrupted by spatially correlated noise, as typically arises in vendor reconstructed data.
|
||
2689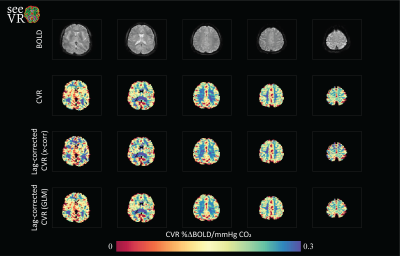 |
53 | seeVR: an open toolbox for analyzing cerebro-vascular reactivity (CVR) data
Alex A Bhogal1
1Radiology, UMC Utrecht, Utrecht, Netherlands
The seeVR toolbox is a series of Matlab functions designed help analyze CVR through rapid implementation of processing pipelines. Depending on the type of stimulus used and the properties of the data acquisition, a number of options are available. All code is open-source and freely downloadable from the seeVR github repository (https://github.com/abhogal-lab/seeVR).
|
||
2690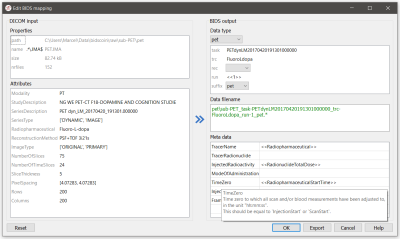 |
54 | BIDScoin: A user-friendly application to convert imaging data to the Brain Imaging Data Structure
Marcel Zwiers1, Cyril Pernet2, Anthony Galassi3, and Robert Oostenveld1,4
1Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 2Neurobiology Research Unit, Copenhagen University Hospital, Copenhagen, Denmark, 3Center for Multimodal Neuroimaging, National Institute of Mental Health, Bethesda, MD, United States, 4NatMEG, Karolinska Institutet, Stockholm, Sweden
Sharing neuroimaging data is an important development that has the potential to be scaled up with the new Brain Imaging Data Structure (BIDS) standard. Existing tools to converting data to BIDS often require programming skills or are tailored to specific institutes, datasets or data formats. Here we introduce BIDScoin, a cross-platform, flexible, free and open-source converter that provides a graphical user interface to help users finding their way in the BIDS standard, and supports plugins to extend its functionality. We show its design and demonstrate how it can be applied to a downloadable tutorial dataset.
|
||
2691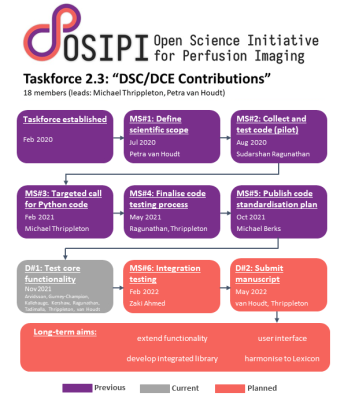 |
55 | Open Science Initiative for Perfusion Imaging (OSIPI): A community-led, open-source code library for analysis of DCE/DSC-MRI
Petra J van Houdt1, Sudarshan Ragunathan2, Michael Berks3, Zaki Ahmed4, Lucy Kershaw5, Oliver Gurney-Champion6, Sirisha Tadimalla7, Jesper Kallehauge8, Jonathan Arvidsson9, Ben Dickie10, Simon Lévy11, Laura Bell12, Steven Sourbron13, and Michael Jonathan Thrippleton14
1Department of Radiation Oncology, the Netherlands Cancer Institute, Amsterdam, Netherlands, 2Neuroimaging Research, Barrow Neurological Institute, Phoenix, AZ, United States, 3Quantitative Biomedical Imaging Laboratory, Division of Cancer Sciences, University of Manchester, Manchester, United Kingdom, 4Radiology, Mayo Clinic, Rochester, MN, United States, 5Edinburgh Imaging and Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, United Kingdom, 6Department of Radiology and Nuclear Medicine, Amsterdam UMC, Amsterdam, Netherlands, 7Institute of Medical Physics, The University of Sydney, Sydney, Australia, 8Department of Oncology, Aarhus University, Aarhus, Denmark, 9Institute of Clinical Sciences, Department of Radiation Sciences, The University of Gothenburg, Gothenburg, Sweden, 10Manchester Academic Health Science Centre, Division of Informatics, Imaging, and Data Science, School of Health Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, United Kingdom, 11Institute of Radiology, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 12Clinical Imaging, Genentech, Inc, South San Francisco, CA, United States, 13Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 14Edinburgh Imaging and Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
A lack of validated, open-source code reduces the reliability of perfusion MRI, resulting in duplicate development. To address this problem, the Open Science Initiative for Perfusion Imaging (OSIPI) established a taskforce to collect, validate and harmonise such code. To date, 74 code contributions have been collected, with 14 of these tested. Source code and tests are published in an open-access repository. The OSIPI DCE/DSC-MRI code collection constitutes a valuable resource for researchers, and will ultimately be developed into a standardised, community-driven open-source code library.
|
||
2692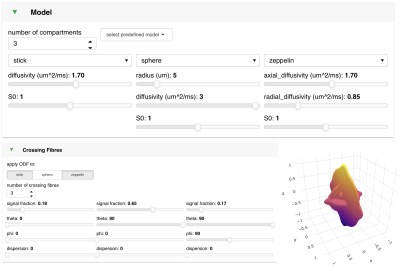 |
56 | Visualising and interacting with diffusion MRI microstructural models using FSL-DIVE
Michiel Cottaar1, Nicole Eichert1, Hossein Rafipoor1, Amy Howard1, Daniel Kor1, Jiewon Kang1, Pavan Chaggar1,2, Ying-Qiu Zheng1, Sean Fitzgibbon1, and Saad Jbabdi1
1University of Oxford, OXFORD, United Kingdom, 2Mathematics, University of Oxford, Oxford, United Kingdom
We introduce FSL-DIVE, an interactive visualisation software for diffusion microstructure modelling. This software allows the user to (i) flexibly set up a multi-compartment microstructure model, including fibre orientation modelling and restricted diffusion, (ii) choose different acquisition parameters, (iii) change the model parameters and analyse the model behaviour through multiple bespoke plots, (iv) save the predicted data (including noise) for any specified acquisition scheme. We hope this tool may be useful for teaching diffusion microstructure modelling, for developing new models, or for getting a better understanding of existing models.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.