Digital Poster
Cardiac Anatomy & Tissue Characterization V
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1754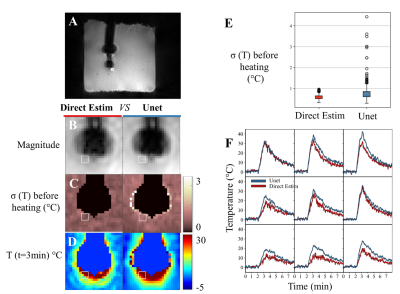 |
24 | Deep artifact suppression for interventional cardiac MR-thermometry
Olivier Jaubert1, Valery Ozenne2,3,4,5, Maxime Yon2,3,4, Javier Montalt-Tordera1, Jennifer Steeden1, Vivek Muthurangu1, and Bruno Quesson2,3,4
1Institute of Cardiovascular Science, University College London, London, United Kingdom, 2Electrophysiology and Heart Modeling Institute, Foundation Bordeaux Université, IHU Lyric, Bordeaux, France, 3Centre de recherche Cardio-Thoracique de Bordeaux, Universite Bordeaux, Bordeaux, France, 4Centre de recherche Cardio-Thoracique de Bordeaux, INSERM, Bordeaux, France, 5Centre de Résonance Magnétique des Systèmes Biologiques, Bordeaux, France A novel thermometry acquisition and a fast deep learning based image reconstruction were combined for cardiac interventional thermometry at high spatial (0.86×0.86mm2) and temporal (0.97s) resolutions, robust to motion and susceptibility artefact and independent of external ECG-gating. The method was tested in phantom and in-vivo in a sheep. The proposed deep learning method outperformed the state-of-the-art algorithm in terms of SNR and paves the way for clinical studies. |
||
1755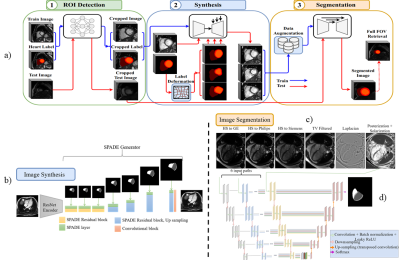 |
25 | Late feature fusion and GAN-based augmentation for generalizable cardiac MRI segmentation
Yasmina Al Khalil1, Sina Amirrajab1, Cristian Lorenz2, Jürgen Weese2, Josien Pluim1, and Marcel Breeuwer1,3
1Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3Philips Healthcare, MR R&D - Clinical Science, Best, Netherlands
While recent deep-learning-based approaches in automatic cardiac magnetic resonance image segmentation have shown great promise to alleviate the need for manual segmentation, most are not applicable to realistic clinical scenarios. This is largely due to training on mainly homogeneous datasets, without variation in acquisition parameters and pathology. In this work, we develop a model applicable in multi-center, multi-disease, and multi-view settings, where we combine heart region detection, augmentation through synthesis and multi-fusion segmentation to address various aspects of segmenting heterogeneous cardiac data. Our experiments demonstrate competitive results in both short-axis and long-axis MR images, without physically acquiring more training data.
|
||
1756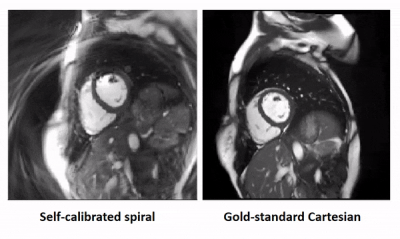 |
26 | Self-calibrated through-time spiral GRAPPA for real-time, free-breathing evaluation of left ventricular function
Dominique Franson1, James Ahad1, Yuchi Liu2, Alexander Fyrdahl2, and Nicole Seiberlich2
1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2University of Michigan, Ann Arbor, MI, United States
Through-time non-Cartesian GRAPPA enables real-time, free-breathing evaluation of the left ventricle, but it requires extra calibration data. This work proposes a self-calibrated spiral GRAPPA method that uses an interleaved acquisition to eliminate the calibration scan. A long spiral trajectory is used to improve robustness to motion. Data were acquired at 12 LV slices with spatiotemporal resolutions of 2.08x2.08 mm2 and 32.72 ms/phase in 71 seconds. In a preliminary study of 6 volunteers comparing the proposed method to a gold-standard, functional values are within the limits of agreement in Bland-Altman analyses and are not statistically significantly different in paired t-tests (p<0.05).
|
||
1757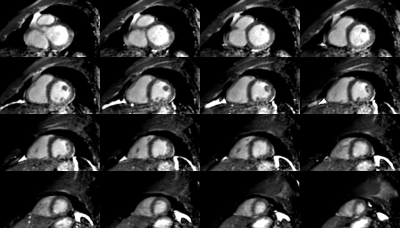 |
27 | Whole-Heart Free-Breathing 3D Cardiac Cine with Fast Respiratory Motion Compensated Reconstruction
Jakob Meineke1, Christophe Schülke1, Kay Nehrke1, and Jochen Keupp1
1Philips Research, Hamburg, Germany
Whole-Heart, Free-Breathing 3D Cardiac Cine MRI with a cartesian, water-selective, balanced SSFP acquisition and spiral-like undersampling is combined with fast, non-rigid motion-compensated reconstruction. It demonstrates the feasibility of routine cardiac cine MRI with minimal planning effort and minimum patient discomfort by avoiding the need for breathholds. The combined acquisition and reconstruction time is below 5 minutes, including computation of deformation vector fields, and minimizes image production delay by starting reconstruction on partially acquired data.
|
||
1758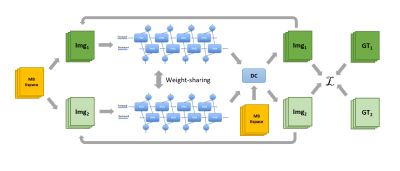 |
28 | Accelerated Multiband Cine Cardiac Magnetic Resonance Imaging using Deep Learning
Xiao Chen1, Yuan Zheng2, Eric Z Chen1, Zhongqi Zhang2, Yu Ding2, Jian Xu2, Terrence Chen1, and Shanhui Sun1
1United Imaging Intelligence, Cambridge, MA, United States, 2UIH America, Inc., Houston, TX, United States Cine CMR is a routinely performed technique for heart anatomy and function analysis. In clinics, repeated breathholdings are performed to acquire multiple 2D slices to cover the whole heart, which further exacerbates the discomfort and challenges for young and severe-diseased patients. In this study, we halved the breathholding time for cine CMR using MB technique. Multiple sampling patterns were designed and studied for the best reconstruction quality. A NN leveraging the Siamese structure was designed to reconstruct the accelerated MB data and achieved great image quality. |
||
1759 |
29 | Transfer Learning in Ultrahigh Field (7T) Cardiac MRI – Automatic Left Ventricular Segmentation in a Porcine Animal Model
Alena Kollmann1, David Lohr1, Markus Ankenbrand2, Maya Bille1, Maxim Terekhov1, Michael Hock1, Ibrahim Elabyad1, Theresa Reiter1,3, Florian Schnitter3, Wolfgang Bauer1,3, Ulrich Hofmann3, and Laura Schreiber1
1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), University Hospital Wuerzburg, Wuerzburg, Germany, 2Center for Computational and Theoretical Biology (CCTB), University of Wuerzburg, Wuerzburg, Germany, 3Department of Internal Medicine I, University Hospital Wuerzburg, Wuerzburg, Germany
Cardiac magnetic resonance (CMR) is considered the gold standard for evaluating cardiac function. Tools for automatic segmentation already exist in the clinical context. To bring the benefits of automatic segmentation to preclinical research, we use a deep learning based model. We demonstrate that for training small data sets with parameters deviating from the clinical situation (high resolution images of porcine hearts acquired at 7T in this case) are sufficient to achieve good correlation with manual segmentation. We obtain DICE-scores of 0.87 (LV) and 0.85 (myocardium) and find high agreement of the calculated functional parameters (Pearson‘s r between 0.95 and 0.99).
|
||
1760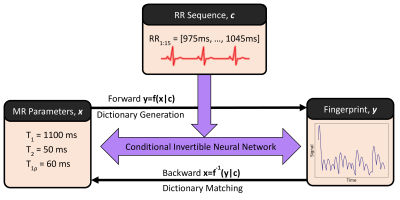 |
30 | Dictionary Generation and Matching with Conditional Invertible Neural Networks for Cardiac MR Fingerprinting
Thomas James Fletcher1, Carlos Velasco1, Gastão Cruz1, Alina Schneider1, René Michael Botnar1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Dictionary generation and pattern matching are two important bottlenecks in cardiac MRF. Dictionaries must be recalculated for each new scan as they depend on the subject’s heart rate variability and both dictionaries and the length of pattern matching grow exponentially with the number of parameters being considered. We propose a conditional invertible neural network capable of both dictionary generation and parameter estimation for T1, T2 and T1ρ cardiac MRF. The network achieves excellent results on EPG-generated data (inner product >0.999, parameters’ relative error <1.5%) and good results for in-vivo data (mean relative errors for myocardium ranging from 2.2% to 15.1%).
|
||
1761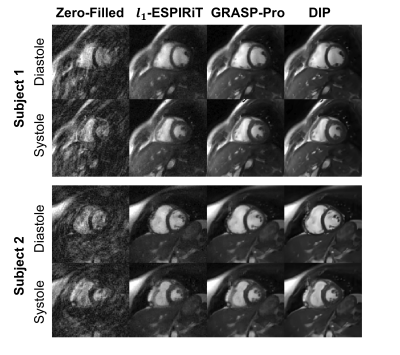 |
31 | Real-Time Spiral bSSFP Functional Cardiac MRI on a 0.55T Scanner Using a Deep Image Prior Reconstruction with GROG
Jesse Ian Hamilton1,2 and Nicole Seiberlich1,2
1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
This work introduces a physics-based deep learning reconstruction that can enable real-time (free-breathing and ungated) spiral 2D bSSFP functional cardiac MRI on a 0.55T scanner. The proposed method extends the concept of a deep image prior and does not require prior training on reference data. In addition, GROG is used to reduce the computational complexity of the forward model calculation by shifting spiral k-space data to their nearest Cartesian grid points, allowing use of FFT rather than NUFFT operations. Results are demonstrated in healthy subjects with functional measurements compared to gold standard acquisitions at 1.5T.
|
||
1762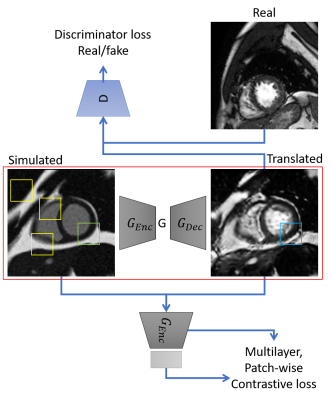 |
32 | sim2real: Cardiac MR image simulation-to-real translation via unsupervised GANs
Sina Amirrajab1, Yasmina Al Khalil1, Cristian Lorenz2, Jürgen Weese2, Josien Pluim1, and Marcel Breeuwer1,3
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands
There has been considerable interest in the MR physics-based simulation of a database of virtual cardiac MR images for the development of deep-learning analysis networks. However, the employment of such a database is limited or shows suboptimal performance due to the realism gap, missing textures, and the simplified appearance of simulated images. In this work we 1) provide image simulation on virtual XCAT subjects with varying anatomies, and 2) propose sim2real translation network to improve image realism. Our usability experiments suggest that sim2real data exhibits a good potential to augment training data and boost the performance of a segmentation algorithm.
|
||
1763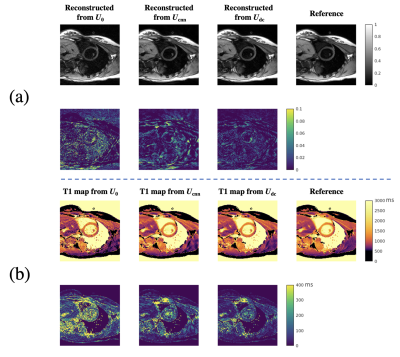 |
33 | Deep subspace transfer learning for SMS image reconstruction from single-slice training: Application to CMR Multitasking
Zihao Chen1,2, Hsu-Lei Lee1, Xianglun Mao1, Tianle Cao1,2, Yibin Xie1, Debiao Li1,2, and Anthony Christodoulou1,2
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States
CMR multitasking is promising for quantitative cardiac imaging, and can achieve fast three-slice quantification when combined with simultaneous multi-slice (SMS) acquisition. However, slow non-Cartesian iterative reconstruction is a barrier to clinical adoption. Deep learning can accelerate reconstruction, but the SMS training data are currently limited. Here we propose a data-consistent deep subspace transfer learning strategy that trains on single-slice T1 CMR multitasking data but is applied to SMS-encoded T1 CMR multitasking image reconstruction. The proposed strategy is >40x faster than the conventional SMS reconstruction, resulting in an equally better image quality and comparably precise T1 as in single-slice reconstruction.
|
||
1764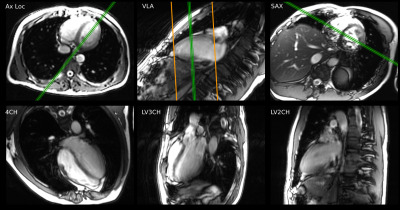 |
34 | Prospective Evaluation of Machine Learning Prescription of Cardiac MRI Planes in Children with Congenital Heart Disease
Cheng William Hong1, Naeim Bahrami2, Vipul R Sheth1, and Shreyas Vasanawala1
1Department of Radiology, Stanford University, Palo Alto, CA, United States, 2GE Healthcare, Menlo Park, CA, United States
Cardiac MRI plays an important role in defining cardiac anatomy and function in children with heart disease. Prescription of diagnostic planes in cardiac MRI requires specialized training, limiting availability. Machine-learning based prescription has the potential to allow rapid, consistent acquisition of these planes independent of operator, but performance in congenital heart disease is unknown. This study evaluates the utility of such a system which demonstrates performance in children with mild to moderate heart disease similar to healthy adult volunteers. As expected, the system does not generate acceptable results in severe intracardiac anomalies.
|
||
1765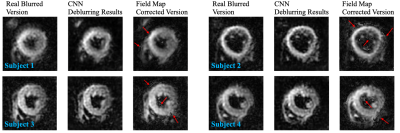 |
35 | CNN-based Off-resonance Correction in Cardiac Spiral MRI
Shishuai Wang1, Pedro F Ferreira2, Zohya Khalique2, Margarita Gorodezky2, Malte Roehl2, Jialin Pu3, Dudley J Pennell2, Sonia Nielles-Vallespin2, and Andrew D Scott2
1Department of Physics, Imperial College London, London, United Kingdom, 2Cardiovascular Magnetic Resonance Unit, Royal Brompton Hospital and National Heart and Lung Institute, Imperial College London, London, United Kingdom, 3College of Information And Communication Engineering, Harbin Engineering University, Harbin, China
Spiral trajectories are time efficient but are susceptible to off-resonance artefacts. Many approaches to off-resonance correction require a high-quality B0 field map. Here we train a convolutional neural network to remove the off-resonance artefacts using simulated cardiac spiral MRI and validate our methods by applying this network to diffusion tensor cardiovascular MR (DTCMR) data acquired with spiral trajectories.
|
||
1766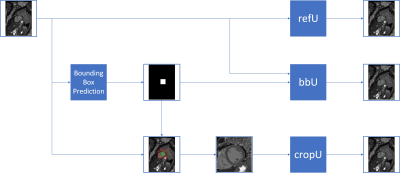 |
36 | Improving U-Net based segmentation in cardiac parametrical T1 mapping by incorporating bounding box information Video Permission Withheld
Darian Viezzer1,2, Thomas Hadler1,2, Edyta Blaszczyk1,2, Maximilian Fenski1,3, Jan Gröschel1,2, Steffen Lange4, and Jeanette Schulz-Menger1,2,3
1Charité Universitätsmedizin Berlin, Working Group on Cardiovascular Magnetic Resonance, Experimental and Clinical Research Center, a joint cooperation between the Charité Universitätsmedizin Berlin and the Max-Delbrück-Center for Molecular Medicine, Berlin, Germany, 2DZHK (German Centre for Cardiovascular Research), partner site Berlin, Berlin, Germany, 3Department of Cardiology and Nephrology, Helios Hospital Berlin-Buch, Berlin, Germany, 4Faculty for Computer Sciences, Hochschule Darmstadt (University of Applied Sciences), Darmstadt, Germany
Parametric mapping images contain to a large extent irrelevant background information. In order to hide some of it, we incorporated bounding box information into a U-net based segmentation network. Our dataset consisted of 845 training, 102 validation and 146 test T1 maps of native and post-contrast myocardium from different clinical studies, including healthy volunteers and patients with inflammatory heart disease, muscular dystrophies or chronic myocardial infarction. While cropping the image input improved the segmentation itself, a second input of the bounding box mask reduced the mean absolute and mean squared T1 deviation, which is clinically preferred.
|
||
1767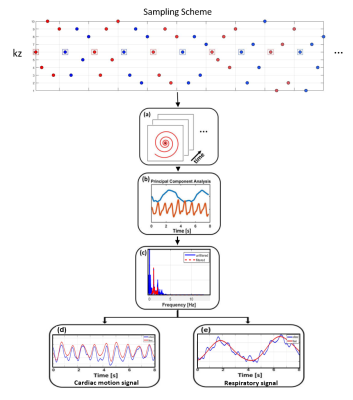 |
37 | Rapid Free-breathing 3D SPirAl Respiratory and Cardiac Self-gated (SPARCS) Cine Acquisition Using an Undersampled Stack-of-Spirals
Xitong Wang1, Junyu Wang1, Ruixi Zhou1,2, and Michael Salerno1,3,4,5
1Department of Biomedical Engineering, University of Virginia, Charottesville, VA, United States, 2School of Artificial Intelligence, Beijing University of Posts and Telecommunications, Beijing, China, 3Department of Medicine, Stanford University, Palo Alto, CA, United States, 4Department of Medicine, University of Virginia, Charottesville, VA, United States, 5Department of Radiology, University of Virginia, Charottesville, VA, United States Cardiac cine images are the gold standard techniques for analyzing cardiac function. Clinical breath-held ECG-gated Cartesian cine imaging requires 10-12 breath-holds to cover the left ventricle. Our group has developed a continuous-acquisition respiratory and cardiac self-gated cine sequence (SPARCS) for free-breathing cardiac function acquisition using 2D spiral acquisition can suffer from through-plane motion. Thus, we develop a rapid 3D self-gated cine technique using a variable density undersampled stack of spiral gradient echo sequence to provide 3D coverage of the ventricle enabling thinner slice acquisition and improved robustness to through-plane motion. |
||
1768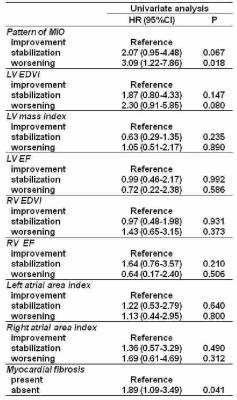 |
38 | Changes in CMR Parameters and Prediction of Cardiac Complications in Thalassemia Major: Fibrosis Tells Us More than Iron.
Antonella Meloni1, Laura Pistoia1, Pietro Giuliano2, Nicola Giunta2, Nicolò Schicchi3, Emanuele Grassedonio4, Stefania Renne5, Vincenzo Positano1, Lorella Pitrolo6, Maria Grazia Roberti7, Paola Maria Grazia Sanna8, Filippo Cademartiri1, and Alessia Pepe1
1Fondazione G. Monasterio CNR-Regione Toscana, Pisa, Italy, 2"ARNAS" Civico, Di Cristina Benfratelli, Palermo, Italy, 3Azienda Ospedaliero-Universitaria Ospedali Riuniti "Umberto I-Lancisi-Salesi", Ancona, Italy, 4Policlinico "Paolo Giaccone", Palermo, Italy, 5Presidio Ospedaliero “Giovanni Paolo II”, Lamezia Terme (CZ), Italy, 6Ospedale "V. Cervello", Palermo, Italy, 7Azienda Ospedaliero-Universitaria OO.RR. Foggia, Foggia, Italy, 8Azienda Ospedaliero-Universitaria di Sassari, Sassari, Italy
Seven-hundred and nine patients with thalassemia major who performed a baseline and a 1st follow-up CMR scan after 18 months were followed prospectively in order to evaluate the predictive value of changes in CMR parameters (myocardial iron, biventricular function, and replacement myocardial fibrosis) for cardiac complications. During a mean follow-up of 89.4±33.3 months, cardiac events (heart failure, arrhythmias, and pulmonary hypertension) were recorded in 7.1% of patients. In the univariate Cox regression analysis, cardiac iron clearance and replacement myocardial fibrosis were identified as univariate prognosticators but in the multivariate analysis only myocardial fibrosis remained an independent predictor factor.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.