Digital Poster
Quality Assessment & Data Harmonization I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1788 |
58 | Predicting artifacts in maximum intensity projections of high b-value DWI of the breast using neural networks.
Andrzej Liebert1, Lorenz Kapsner1, Lukas Folle2, Hannes Schreiter1,2, Badhan Kumar Das1,2, Sabine Ohlmeyer1, Andreas Maier2, Evelyn Wenkel1, Frederik B. Laun1, Michael Uder1, and Sebastian Bickelhaupt1
1Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 2Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander University Erlangen Nuremberg, Erlangen, Germany DWI acquisitions in MRI are prone to artifacts caused e.g. by insufficient fat saturation and can significantly impede the diagnostic assessment. We investigated the ability to predict the occurrence of these artifacts in maximum intensity projections (MIPs) of high b-value DWI using a neuronal network that analyses T2w images, which were acquired prior to the DWI sequence. An AuRoC of 0.83 with sensitivity of 0.82 and specificity of 0.61 was achieved. Investigation of the regions of the T2w images most important for the decision of the neural network showed a good correlation with artifact-affected areas in the DWI MIP images. |
||
1789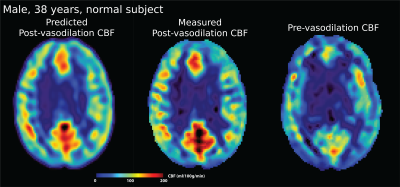 |
59 | Predicting Perfusion Augmentation Using Deep Learning without Vasodilators
Moss Zhao1, Ramy Hussein1, Michael Moseley1, and Greg Zaharchuk1
1Department of Radiology, Stanford University, Stanford, CA, United States
We present a deep learning technique to predict cerebral perfusion after vasodilation challenges. A 3D convolutional neural network (CNN)-based encoder-decoder architecture was constructed to transform ASL perfusion images acquired pre-vasodilation into post-vasodilation images using an improved attention-gated 3D U-Net. Results showed that the prediction and ground truth were not significantly different. This technique will enable a drug-free MR procedure to study the hemodynamic of patients with high risk cerebrovascular diseases.
|
||
1790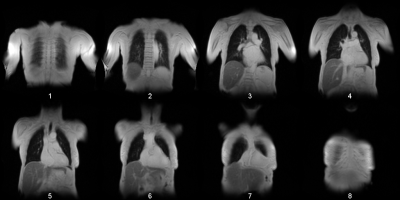 |
60 | Convolutional Neuronal Network Inception-v3 detects Partial Volume Artifacts on 2D MR-Images of the Lung for Automated Quality Control
Andreas Voskrebenzev1,2, Cristian Crisosto1,2, Maximilian Zubke1,2, Frank Wacker1,2, and Jens Vogel-Claussen1,2
1Institute of Diagnostic and Interventional Radiology, Hannover Medical School, Hannover, Germany, 2German Center for Lung Research (DZL), Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), Hannover, Germany
The partial volume effect (PVE) is an often-observed artifact in MR imaging. Especially images with a low spatial resolution, will show an averaged voxel signal of multiple tissue components. These artifacts can be so substantial that a further image analysis can be omitted. This is e.g. the case for phase-resolved functional lung imaging (PREFUL), which is based on the 2D acquisition of coronal image-time-series to assess ventilation and perfusion dynamics. In this study the pretrained convolutional neural network Inception-v3 was trained via transfer-learning to detect images, which show substantial PVE with a classification accuracy of 91%.
|
||
1791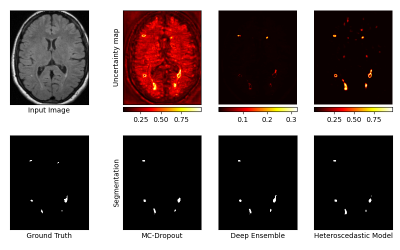 |
61 | Multi-Scale Evaluation of Uncertainty Quantification Techniques for Deep Learning based MRI Segmentation
Benjamin Lambert1,2, Florence Forbes3, Alan Tucholka2, Senan Doyle2, and Michel Dojat1
1Univ. Grenoble Alpes, Inserm, U1216, Grenoble Institut Neurosciences, GIN, 38000, Grenoble, France, 2Pixyl, Research and Development Laboratory, 38000, Grenoble, France, 3Univ. Grenoble Alpes, Inria, CNRS, Grenoble INP, LJK, 38000, Grenoble, France
Efforts are required to design Deep Learning models that are not only powerful, but also capable of expressing the certainty of their predictions.
We evaluate 3 state-of-the-art techniques for uncertainty quantification : Monte-Carlo Dropout, Deep Ensemble, and Heteroscedastic models. Evaluation is illustrated on a task of automatic segmentation of White-Matter Hyperintensities in T2-weighted FLAIR MRI sequences of Multiple-Sclerosis patients. Analysis is performed at 3 different scales : the voxel, the lesion, and the whole image. Results indicate the superiority of the Heteroscedastic approach, which ranked first in both the uncertainty and segmentation tasks. |
||
1792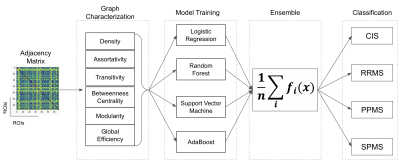 |
62 | Grey Matter Connectome Data Analysis and Classification of Multiple Sclerosis Clinical Profiles Video Not Available
Berardino Barile1, Claudio Stamile2, Sabine Van Huffel3, and Dominique Sappey-Marinier4
1CREATIS (UMR5220 CNRS & U1294 INSERM, Université Claude Bernard-Lyon1 & INSA-Lyon, France, Villeurbanne, France, 2R&D Department CGnal Milan, Italy, Villeurbanne, France, 3KU Leuven, Leuven, Belgium, 4CREATIS (UMR5220 CNRS & U1294 INSERM), Université Claude Bernard-Lyon1 & INSA-Lyon, France, Villeurbanne, France
Multiple Sclerosis (MS) is an autoimmune inflammatory disease characterized by white matter (WM) lesions and gray matter (GM) degeneration leading to cognitive and physical impairments. Recently, brain connectivity techniques were developed to better characterize such brain damages in association with the clinical profiles of MS patients. In this study, we proposed a complete automated pipeline to first extract global graph metrics from the morphological brain connectivity based on GM tissue thickness and second to train a higher-level ensemble of four machine learning models for the classification of MS clinical profiles using only the T1-weighted image modality.
|
||
1793 |
63 | Analysing The Role Of Model Uncertainty in Flourine-19 MRI using Markov Chain Monte Carlo methods
Masoumeh Javanbakhat1, Ludger Starke2, Sonia Waiczies2, and Christoph Lippert1
1Digital Health-Machine Learning Group, Hasso Plattner Institute, Potsdam, Germany, 2Berlin Ultrahigh Field Facility, Max Delbrück Centre for Moleculare Medicine in the Helmholtz Association, Berlin, Germany
Deep learning (DL) has achieved state of the art results in semantic segmentation of numerous medical imaging applications. Despite promising results deep learning models tend to produce point estimates as outputs which leads to overconfident, miscalibrated predictions. These overconfident predictions are specifically problematic in medical applications. Hence, providing a measure of a system’s confidence to identify untrustworthy predictions is essential to guide clinical decisions. Here we propose a 3D Bayesian segmentation model to provide uncertainty estimation for the Fluorine-19 MRI dataset based on Stochastic Gradient Markov Chain Monte Carlo methods.
|
||
1794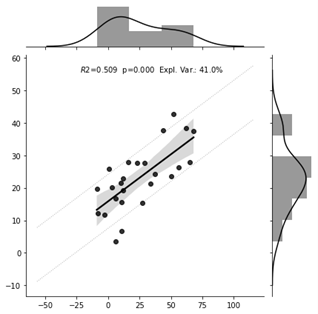 |
64 | Resting-state functional connectivity predicts subsequent pain-related threat learning
Balint Kincses1,2, Katarina Forkmann1, Katharina Schmidt1, Ulrike Bingel1, and Tamas Spisak2
1Bingel-laboratory, Department of Neurology, University Hospital Essen, Essen, Germany, 2Laboratory of Predictive NeuroImaging, Institute for Diagnostic and Interventional Radiology and Neuroradiology, University Hospital Essen, Essen, Germany
Fear conditioning has a role in anxiety-disorders and the neurobiological correlates of it are not yet well understood. Therefore, we trained a machine learning predictive model on individual functional resting state connectivity data to predict the emotional aspects of fear conditioning. The model was found to predict individual pain-related threat learning measured by the change of valence with an explained variance of 24%-41%. These results highlight the potential of machine learning to enhance our understanding of fear conditioning.
|
||
1795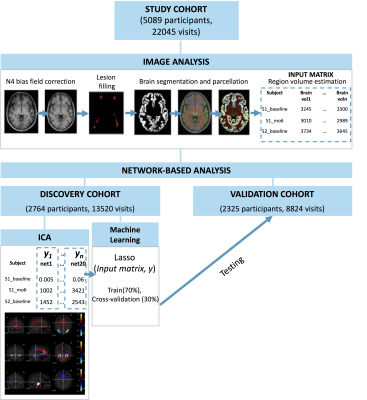 |
65 | Developing pattern recognition models to extract longitudinal network-based measures at an individual level Video Permission Withheld
Elisa Colato1, Claudia AM Wheeler-Kingshott1,2,3, Douglas L Arnold4, Frederik Barkhof1,5,6,7, Olga Ciccarelli1,5, Declan Chard1,5, and Arman Eshaghi1,8
1Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London (UCL), London, United Kingdom, 2Brain MRI 3T Research Centre, C. Mondino National Neurological Institute, Pavia, Italy, 3Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 4McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, QC, Canada, 5Institute for Health Research (NIHR), University College London Hospitals (UCLH) Biomedical Research Centre (BRC), London, United Kingdom, 6Department of Radiology and Nuclear Medici, VU medical centre, Amsterdam, Netherlands, 7Centre for Medical Image Computing (CMIC), Department of Medical Physics and Biomedical Engineering, University College London (UCL), London, United Kingdom, 8Centre for Medical Image Computing (CMIC), Department of Computer Science, University College London (UCL), London, United Kingdom
Network-based measures can outperform regional and whole-brain grey matter (GM) measures in explaining clinical disability in several neurodegenerative disorders. However, network measures are mostly estimated at the group level and require a re-estimation of model parameters when applied to new participants. Here, we introduce a new longitudinal network analysis paradigm to extract longitudinal ICA-like components at an individual level from a discovery cohort, applied machine learning to obtain individual-level network-based measure for a validation cohort, and used them to explain clinical disability in multiple sclerosis.
|
||
1796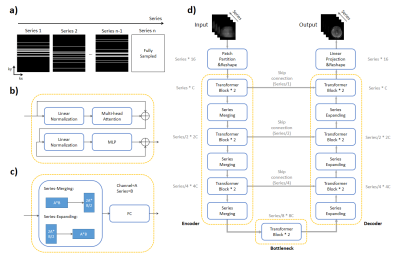 |
66 | Deeping-Learning based acceleration for Serial MR imaging using Transformer (SMR-Transformer): Comparison with CNN on Three Datasets
Chuyu Liu1, Shuo Chen1, Yibing Chen2, Rui Li1, and Xiaolei Song1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Xi’an Key Lab of Radiomics and Intelligent Perception, School of Information Sciences and Technology, Northwest University, Xi'an, China
Serial image acquisition is always required for quantitative and dynamic MRI, in which the spatial and temporal resolution as well as the contrast is restricted by acquisition speed. Herein we employed a novel transformer based deep-learning method for reconstruction of highly under-sampled MR serial images. The network architecture and input were designed to take advantages of Transformer’s capacity on global information learning along the redundant series dimension. SMR-transformer was compared with 3D Res-U-Net, on a self-acquired CEST dataset, a cardiac dataset from OCMR and a diffusion dataset from HCP. Both images and quantitative comparison indicated SMR-Transformer allowed fast serial MRI.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.