Digital Poster
Quality Assessment & Data Harmonization II
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1870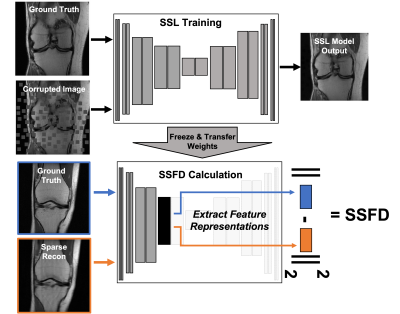 |
50 | SSFD: Self-Supervised Feature Distance Outperforms Conventional MR Image Reconstruction Quality Metrics
Philip M. Adamson1, Jeffrey Dominic1, Arjun Desai1, Christian Bluethgen2, Jeff P. Wood3, Ali B. Syed2, Robert Boutin2, Kathryn J. Stevens2, Daniel Spielman2, Shreyas Vasanawala2, John M. Pauly1, Akshay S. Chaudhari2, and Beliz Gunel1
1Department of Electrical Engineering, Stanford University, Palo Alto, CA, United States, 2Department of Radiology, Stanford University, Palo Alto, CA, United States, 3Austin Radiological Association, Austin, TX, United States
Evaluation of accelerated magnetic resonance imaging (MRI) reconstruction methods is imperfect due to the discordance between quantitative image quality metrics (IQMs) and radiologist-perceived image quality. Self-supervised learning (SSL) is a deep learning (DL) method that has become a popular pre-training tool due to its ability to capture generalizable and domain-specific feature representations of the underlying data without the need for labels. In this study, we derive a data-driven self-supervised feature distance (SSFD) IQM to assess MR image reconstruction quality. We demonstrate that SSFD is more highly correlated to three radiologist’s perceived image quality on DL-based sparse reconstructions than conventional IQMs.
|
||
1871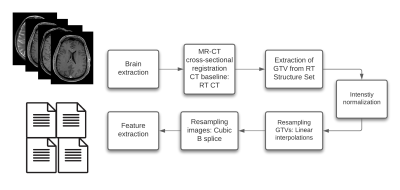 |
51 | Impact Of MR Intensity Normalization For Different MR Sequences In MRI Based Radiomics Studies
Patrick Salome1,2,3,4, Francesco Sforazzini1,2,3,4, Andreas Kudak3,5,6, Matthias Dostal3,5,6, Nina Bougatf3,5,6, Jürgen Debus3,4,5,7, Maximilian Knoll1,3,4,5, and Amir Abdollahi1,3,4,5
1CCU Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Medical Faculty, Heidelberg University Hospital, Heidelberg, Germany, 3Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg, Germany, 4German Cancer Consortium (DKTK) Core Center, Heidelberg, Germany, 5Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany, 6CCU Radiation Therapy, German Cancer Research Center (DKFZ), Heidelberg, Germany, 7National Center for Tumor Diseases (NCT), Heidelberg, Germany
MR-based radiomics prognostic signatures have great clinical potential but are currently hindered due to a lack of standardisation in the radiomics workflow. This study focuses on a crucial step of these workflows, i.e. intensity normalisation, while presenting a methodology that allows for determining the most suitable MR intensity normalization method for a specific entity.
|
||
1872 |
52 | Cortical thickness predicts pain sensitivity in a multi-centre cohort: a machine learning approach
Raviteja Kotikalapudi1,2, Balint Kincses1,2,3, Kevin Hoffschlag1, Matthias Zunhammer2, Tobias Schmidt-Wilcke4,5, Zsigmond T Kincses3, Ulrike Bingel2, and Tamas Spisak1,2
1Laboratory of Predictive NeuroImaging, Institute for Diagnostic and Interventional Radiology and Neuroradiology, University Hospital Essen, Essen, Germany, 2The Bingel Laboratory, Translational Pain Research Unit, University Hospital Essen, Essen, Germany, 3Department of Neurology, University of Szeged, Szeged, Hungary, 4Institute for Clinical Neurosciences and Medical Psychology, Heinrich Heine University, Dusseldorf, Germany, 5Neurocentre, District Hospital Mainkofen, Mainkofen, Deggendorf, Germany
Individual sensitivity to pain is both a precursor and a symptom of many clinical pain conditions. A pain predictive model would have potential applications in objectively characterizing pain in acute and chronic pain individuals. Here, we developed a cortical thickness-based predictive model of pain sensitivity using a machine learning approach and multi-centre T1-weighted MRI and quantitative pain threshold data. We found that our model significantly predicts pain sensitivity, that was measured through heat, cold and mechanical stimuli. Furthermore, the predictions were exclusively driven by cortical thickness and not confounded by variables of demographic and psychological value.
|
||
1873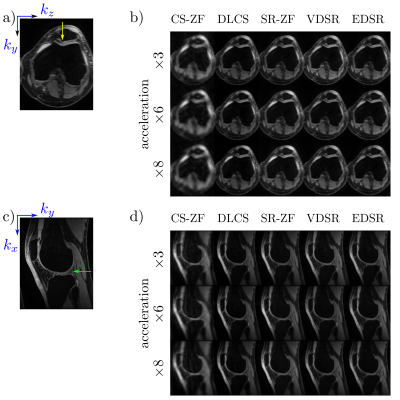 |
53 | Benchmarking Accelerated MRI: A Head-to-Head Comparison of Deep Learning Reconstruction and Super-Resolution Techniques
Eric K. Gibbons1, Zhongnan Fang2, Arjun D. Desai3, Christopher M. Sandino3, Garry E. Gold4, Brian A. Hargreaves4, and Akshay S. Chaudhari4
1Department of Electrical and Computer Engineering, Weber State University, Ogden, UT, United States, 2Lvis Corporation, Palo Alto, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 4Department of Radiology, Stanford University, Stanford, CA, United States
Deep-learning (DL) can be used to extend compressed sensing (CS) to learn the regularization function in a data-driven manner. In contrast, super resolution (SR) algorithms have been used to transform rapidly-acquired low-resolution images into higher-resolution images. This work compares DL-CS with DL-SR for accelerated MRI on a test dataset of 50 patients with conventional image quality metrics and clinically-relevant quantitative T2 relaxation measurements. We demonstrate that DLCS approaches outperform DLSR approaches for accelerated MRI.
|
||
1874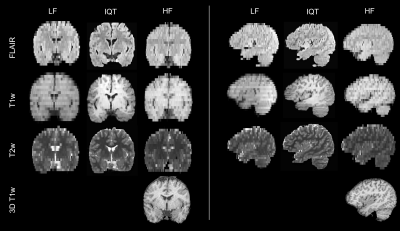 |
54 | Image Quality Transfer improves the potential clinical value of low-field MRI
Matteo Figini1,2, Hongxiang Lin1,2,3, Felice D'Arco4, Godwin Inalegwu Ogbole5, Maria Camilla Rossi Espagnet6, Olalekan Ibukun Oyinloye7, Joseph O Yaria8, Donald Amasike Nzeh7, Mojisola Omolola Atalabi9, Lisa Ronan1,2, David W Carmichael10,11, Judith Helen Cross4,11, Ikeoluwa A Lagunju12, Delmiro Fernandez-Reyes2,12, and Daniel C Alexander1,2
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Computer Science, University College London, London, United Kingdom, 3Research Center for Healthcare Data Science, Zhejiang Lab, Hangzhou, China, 4Great Ormond Street Hospital for Children, London, United Kingdom, 5Radiology, College of Medicine, University of Ibadan, Ibadan, Nigeria, 6Neuroradiology, Sapienza University, Rome, Italy, 7Radiology, University of Ilorin Teaching Hospital, Ilorin, Nigeria, 8Neurology, University College Hospital Ibadan, Ibadan, Nigeria, 9Radiology, University College Hospital Ibadan, Ibadan, Nigeria, 10School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 11UCL Great Ormond Street Institute of Child Health, London, United Kingdom, 12Paediatrics, College of Medicine, University of Ibadan, Ibadan, Nigeria
We applied Image Quality Transfer to enhance the contrast and resolution in the slice direction of low-field structural MRI, using a deep learning model trained on simulated images. Six radiologists blindly reviewed the enhanced images compared to low- and high-field images from 12 paediatric patients with epilepsy. Results demonstrated significant improvement of the visualisation of brain structures in sagittal and coronal orientations, and marginal improvement of the contrast between grey and white matter. If these promising results are confirmed in a larger study and in lesions, IQT could be an important tool to enhance the diagnostic power of low-field MRI.
|
||
1875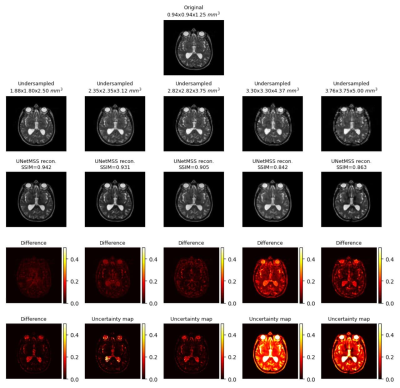 |
55 | Uncertainty quantification for ground-truth free evaluation of deep learning reconstructions
Soumick Chatterjee1,2,3, Alessandro Sciarra1,4, Max Dünnwald3,4, Anitha Bhat Talagini Ashoka3, Mayura Gurjar Cheepinahalli Vasudeva3, Shudarsan Saravanan3, Venkatesh Thirugnana Sambandham3, Steffen Oeltze-Jafra4,5, Oliver Speck1,5,6,7, and Andreas Nürnberger2,3,5
1Department of Biomedical Magnetic Resonance, Otto von Guericke University Magdeburg, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University Magdeburg, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University Magdeburg, Magdeburg, Germany, 4MedDigit, Department of Neurology, Medical Faculty, University Hospital, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6German Centre for NeurodegenerativeDiseases, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
Many deep learning-based techniques have been proposed in recent years to reconstruct undersampled MRI – showing their potential for shortening the acquisition time. Before using them in actual practice, they are usually evaluated by comparing their results against the available ground-truth – which is not available during real applications. This research shows the potential of using uncertainty estimation to evaluate the reconstructions without using any ground-truth images. The method has been evaluated for the task of super-resolution MRI, for acceleration factors ranging from two to four – in all three dimensions.
|
||
1876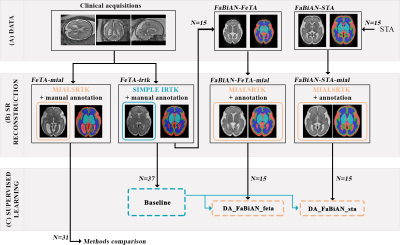 |
56 | On the importance of fetal brain numerical models for domain adaptation strategies in fetal brain MRI tissue segmentation Video Permission Withheld
Priscille de Dumast1,2, Hamza Kebiri1,2, Meritxell Bach Cuadra1,2, and Hélène Lajous1,2
1CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland
Manual fetal brain tissue segmentation is needed for training machine learning methods but is a tedious and error-prone task. The generation of synthetic magnetic resonance images can overcome the lack of clinical annotations by supplementing scarce clinical fetal datasets. However, we highlight that the choice of the numerical model from which additional data are derived is key to maximize the segmentation accuracy of clinical data via domain adaptation strategies. We demonstrate that the resort to high-resolution segmented images from real neurotypical and pathological cases enhances the morphological variability compared to an atlas, resulting in improved fetal brain tissue delineation overall.
|
||
1877 |
57 | A deep learning-based quality control system for co-registration of prostate MR images
Mohammed R. S. Sunoqrot1, Kirsten M. Selnæs1,2, Bendik S. Abrahamsen1, Alexandros Patsanis1, Gabriel A. Nketiah1,2, Tone F. Bathen1,2, and Mattijs Elschot1,2
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology (NTNU), Trondheim, Norway, 2Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
Multiparametric MRI (mpMRI) is a valuable tool for the diagnosis of prostate cancer. Computer-aided detection and diagnosis (CAD) systems have the potential to improve robustness and efficiency compared to traditional radiological reading of mpMRI in prostate cancer. Co-registration of diffusion-weighted and T2-weighted images is a crucial step of CAD but is not always flawless. Automated detection of poorly co-registered cases would therefore be a useful supplement. This work shows that a fully automated quality control system for co-registration of prostate MR images based on deep learning has potential for this purpose.
|
||
1878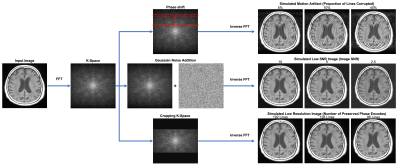 |
58 | MR image enhancement using a multi-task neural network trained using only synthetic data
Kevin Blansit1, Zhehao Hu1, Greg Zaharchuk1, Enhao Gong1, and Keshav Datta1
1Subtle Medical, Menlo Park, CA, United States
Faster MRI scans can be achieved by acquiring low resolution images or low SNR images and enhancing them to standard-of-care using deep learning techniques. However, to achieve clinical diagnostic quality images, this requires a large number of paired clinical datasets to train the model. Here we show that a multi-task deep convolutional neural network (DCNN) trained using only simulated motion artifact, low SNR, and low resolution images is capable of improving the quality of clinically acquired images from motion corrupted and accelerated sequences.
|
||
1879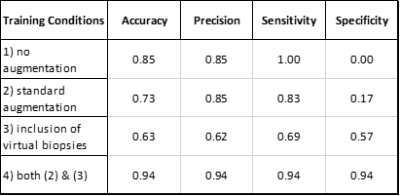 |
59 | The Utility of Virtual Biopsies for Dataset Augmentation as Applied to AI-Based Detection of Tumor Infiltration in Non-Enhancing Brain Lesion
Robert Wujek1, Melissa Prah2, Mona Al-Gizawiy2, and Kathleen Schmainda2
1Graduate School, Medical College of Wisconsin, Wauwatosa, WI, United States, 2Biophysics, Medical College of Wisconsin, Wauwatosa, WI, United States
Delineation of invasive tumor from peritumoral edematous tissue remains a major obstacle to glioma treatment. To address this problem, a neural network was trained to distinguish between these regions using biopsies paired with colocalized MRI inputs. In addition to histologically confirmed biopsies, virtual biopsies sampled from non-contrast enhancing, FLAIR enhancing regions of non-invasive tumors (meningioma, metastasis) were used with an assumed classification of “non-tumor”. The current work is a preliminary assessment of this assumption and it's impact on model performance.
|
||
1880 |
60 | QC of image registration using a DL network trained using only synthetic images
Yiheng Li1
1Subtle Medical, Santa Clara, CA, United States
To perform robust quality control of medical imaging registration, we propose a method that can QC co-registration for multiple organs, without the restriction of the modalities. A rule-based image synthesis pipeline is used to generate random contrasts and shapes as training images. ResNet34 is trained to predict image alignment. Two MRI datasets with the spine or brain as subjects are used as external validation sets. The proposed model trained with synthetic images is validated on either one of the real MRI datasets and outperforms the same model trained on the other MRI dataset, which shows better generalizability.
|
||
1881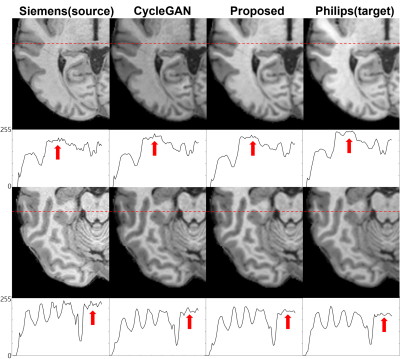 |
61 | Paired CycleGAN-based Cross Vendor Harmonization
Joonhyeok Yoon1, Sooyeon Ji1, Eun-Jung Choi1, Hwihun Jeong1, and Jongho Lee1
1Seoul National University, Seoul, Korea, Republic of
CycleGAN shows good performance with harmonization task. However, generative models have risk of structure modification. we proposed a cross-vendor harmonization model with paired CycleGAN based architecture for both high performance and structural consistency. We acquired 4 in-vivo dataset from Siemens and Philips scanner. For algorithm, we adapted CycleGAN for generation model, and we utilized L1 loss from pix2pix and patchGAN discriminator for structural consistency. Evaluations were performed both quantitatively and qualitatively. To quantitative evaluation, we assessed means of structural similarity index measure (SSIM). Proposed model shows better results compared to CycleGAN architecture.
|
||
1882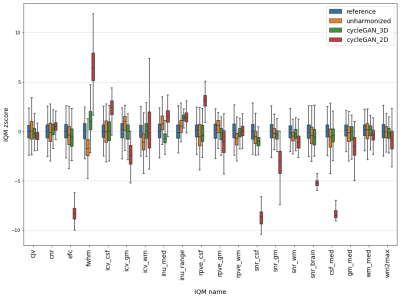 |
62 | Inter-scanner harmonization of T1-weighted brain images using 3D CycleGAN
Vincent Roca1, Grégory Kuchcinski1,2, Morgan Gautherot1, Xavier Leclerc1,2, Jean-Pierre Pruvo1,2, and Renaud Lopes1,2
159000, Univ Lille, UMS 2014 – US 41 – PLBS – Plateformes Lilloises en Biologie & Santé, Lille, France, 259000, Univ Lille, Inserm, Lille Neuroscience & Cognition, Lille, France
In MRI multicentric studies, inter-scanner harmonization is necessary to avoid taking into account variations due to technical differences in the analysis. In this study, we focused on CycleGAN models for 3D T1 weighted brain images harmonization. More precisely, we didn't follow the classical 2D CycleGAN architecure and developped a 3D cycleGAN model. We compared harmonization quality of these two kinds of models using 20 imaging features quantifying T1 signal, contrast between brain structures and segmentation quality. Results illustrate the potential of 3D CycleGAN for better synthesize images in inter-scanner MRI harmonization tasks.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.