Digital Poster
Deep/Machine Learning-Based Image Acquisition & Reconstruction I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
1050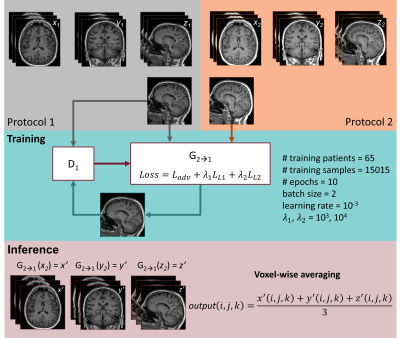 |
57 | Protocol harmonization using a generative adversarial network decreases morphometry variability
Veronica Ravano1,2,3, Jean-François Démonet2, Daniel Damian2, Reto Meuli2, Gian Franco Piredda1,2,3, Till Huelnhagen1,2,3, Bénédicte Maréchal1,2,3, Jean-Philippe Thiran2,3, Tobias Kober1,2,3, and Jonas Richiardi2
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
In radiology, the deployment of automated clinical decision support tools to new institutions is often hindered by inter-site data variability. In MRI, data heterogeneity often arises from differences in acquisition protocols. To overcome this issue, we propose a post-hoc harmonization technique based on generative adversarial networks (GAN). Seventy-seven patients suffering from dementia were scanned with two distinct T1-weighted MP-RAGE protocols. We show that cross-protocol harmonization of brain images using a conditional GAN improves image similarity and reduces the variability of brain morphometry.
|
||
1051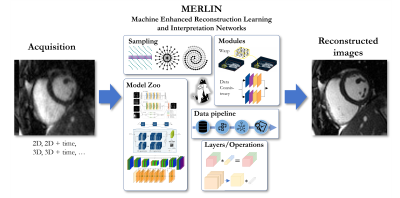 |
58 | Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN)
Kerstin Hammernik1,2 and Thomas Küstner3
1Lab for AI in Medicine, Technical University of Munich, Munich, Germany, 2Department of Computing, Imperial College London, London, United Kingdom, 3Medical Image And Data Analysis (MIDAS.lab), Department of Interventional and Diagnostic Radiology, University Hospital of Tuebingen, Tuebingen, Germany
Machine Learning (ML) methods have evolved tremendously during the last decade. A number of frameworks support the development of new ML methods. However, support for high-dimensional and complex-valued data processing is often limited. Therefore, we propose a Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN) framework that seamlessly integrates with existing ML solutions such as Tensorflow/Keras and Pytorch and complements them by high-dimensional, complex-valued and MR-specific operators and layers. Furthermore, standard data processing pipelines in Python are provided in the context of MRI reconstruction.
|
||
1052 |
59 | A data-driven method for automatic regularization selection in a hybrid DL-SENSE reconstruction
Zahra Hosseini1, Thorsten Feiweier2, John Conklin3, Stephan Kannengiesser2, Marcel Dominik Nickel2, Min Lang3, Azadeh Tabari3, Augusto Lio Concalves Filho3, Wei-Ching Lo4, Maria Gabriela Figueiro Longo3, Michael Lev3, Pamela Schaefer3, Otto Rapalino3, Susie Huang3, Stephen Cauley5, and Bryan Clifford4
1MR R&D Collaboration, Siemens Medical Solutions USA, Atlanta, GA, United States, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 4MR R&D Collaboration, Siemens Medical Solutions USA, Boston, MA, United States, 5Department of Radiology, A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
The integration of deep learning priors into regularized CG-SENSE reconstructions enables high quality MR images to be generated from noisy, undersampled data. The regularization parameter in these methods can be tuned to control the level of denoising, allowing a network to generalize to novel SNR conditions without retraining. However, manual tuning of the regularization parameter can be time consuming. This work presents a data-driven method for automatic regularization selection using commonly acquired noise calibration data. Results indicate the method generalizes across clinically relevant imaging scenarios and provides diagnostically equivalent image quality to that obtained by manual parameter tuning.
|
||
1053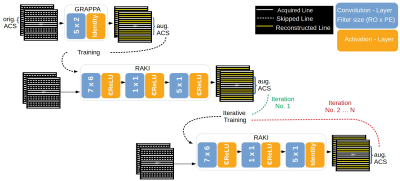 |
60 | Iterative RAKI with complex-valued convolution for improved image reconstruction with limited training samples
Peter Dawood1,2, Martin Blaimer3, Felix Breuer3, Paul R. Burd4, István Homolya5,6, Peter M. Jakob1, and Johannes Oberberger2
1Department of Physics, University of Würzburg, Würzburg, Germany, 2Department of Internal Medicine I, University Hospital Würzburg, Würzburg, Germany, 3Magnetic Resonance and X-ray Imaging Department, Fraunhofer IIS, Fraunhofer Institute for Integrated Circuits IIS, Division Development Center X-Ray Technology, Würzburg, Germany, 4Institute for Theoretical Physics and Astrophysics, University of Würzburg, Würzburg, Germany, 5Brain Imaging Centre, Research Centre for Natural Sciences, Budapest, Hungary, 6Institute of Nuclear Techniques, Budapest University of Technology and Economics, Budapest, Hungary
Recently, the Parallel Imaging method GRAPPA has been generalized by the deep-learning method RAKI, in which Convolutional Neural Networks are used for non-linear k-space interpolation. RAKI uses scan-specific training data, however, due to its increased parameter-space, its reconstruction quality may deteriorate given a limited training-data amount. We evaluate an approach that includes augmented training-data via an initial GRAPPA k-space reconstruction, and weights refinement by iterative training. Thereby, severe residual artefacts are suppressed in RAKI, while preserving its resilience against g-factor noise enhancement in GRAPPA for standard 2D imaging at medium accelerations, for strongly varying contrast between training- and interpolation-data, too.
|
||
1054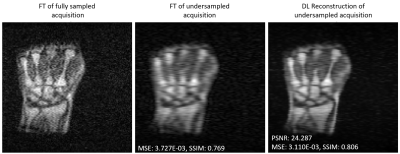 |
61 | Data scarcity mitigation approaches in deep learning reconstruction of undersampled low field MR images
Tobias Senft1, Reina Ayde1, Najat Salameh1, and Mathieu Sarracanie1
1Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland Low magnetic field (LF) MRI is gaining popularity as a flexible and cost-effective complement to conventional MRI. However, LF-MRI suffers from a low signal-to-noise ratio per unit time which calls for signal averaging and hence prolonged acquisition times, challenging the clinical value of LF MRI. In this study, we show that Deep Learning (DL) can reconstruct artifact-free heavily undersampled 2D LF MR images (34% sampling) with great success, both retrospectively and prospectively. Our results also highlight that a transfer learning approach combined with data augmentation improves the overall reconstruction performances, even when only small LF training datasets are available. |
||
1055 |
62 | Concept of a symmetry-guided single-layer neural network for image reconstruction of undersampled radial-MRI k-space data.
Sjoerd Ypma1, Ivo Maatman1, Matthan Caan2, Dimitris Karkalousos2, Marnix Maas1, and Tom Scheenen1
1Radboud UMC, Radiology and Nuclear Medicine, University of Nijmegen, Nijmegen, Netherlands, 2Amsterdam UMC, Biomedical Engineering and Physics, University of Amsterdam, Amsterdam, Netherlands Undersampled k-space data reconstruction results in aliasing artifacts. Compressed sensing theory enables image reconstruction by using a priori knowledge in the form of regularization. Increasingly, Machine Learning methods are used to learn the regularization from data itself, but these methods can result in unstable reconstructions. We propose a translation equivariant single-layer neural network for reconstruction of radially measured k-space data. By exploiting translation symmetry, it can learn from randomly simulated data while still being applicable to in-vivo measurements. We tested robustness to small perturbations and reliability of the reconstruction of unexpected objects. |
||
1056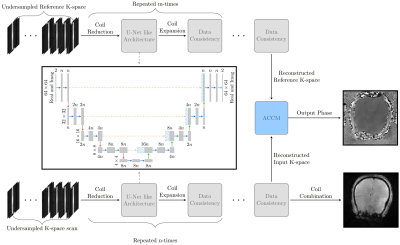 |
63 | Accelerating MR Elastography using Deep Learning-Reconstruction of Undersampled Data
Robin Antony Birkeland Bugge1, Jon Andre Ottesen2, Elies Fuster3, Atle Bjørnerud2, and Kyrre Eeg Emblem1
1Department of Diagnostic Physics, Oslo University Hospital, Oslo, Norway, 2Department of Computational Radiology and Artificial Intelligence, Oslo University Hospital, Oslo, Norway, 3Biomedical Data Science Laboratory, Instituto Universitario de Tecnologías de la Información y Comunicaciones, Universitat Politècnica de València, Valencia, Spain
Problem summary: Brain MR elastography is associated with extended acquisition times, which is alleviated by reduced coverage or resolution. The aim of this project is to utilize deep learning to accelerate MRE. Methods: We employ an MRE for fully sampled acquisition. Undersampled data is simulated by masking phase-encoding steps. A cascaded reconstruction network is used to reconstruct the phase image from undersampled k-space. Results: There are subtle differences between the reconstructed and fully sampled phase images. We observe a non-significant difference for stiffness values in our preliminary results. Conclusions: The method shows promise for accelerating MR elastography data.
|
||
1057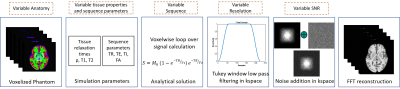 |
64 | Brain MR image super resolution using simulated data to perform in real-world MRI
Aymen Ayaz1, Kirsten Lukassen1, Cristian Lorenz2, Juergen Weese2, and Marcel Breeuwer1,3
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Philips Research Laboratories, Hamburg, Germany, 3MR R&D – Clinical Science, Philips Healthcare, Best, Netherlands
We propose to simulate a large set of anatomically variable voxel-aligned and artifact-free brain MRI data at different resolutions to be used for training deep-learning based Super Resolution (SR) networks. To the best of our knowledge, no such efforts have been made in past regarding use of simulated data to train a SR network. We trained a SR network using such simulated data and tested the performance on real-world MRI data. The trained network could significantly sharpen low-resolution input MR images and clearly outperformed classic image interpolation methods.
|
||
1058 |
65 | Complex graph convolutional neural networks compared to GRAPPA for reconstruction of undersampled non-Cartesian MRSI
Paul Weiser1, Stanislav Motyka2, Wolfgang Bogner2, and Georg Langs1
1Computational Imaging Research Lab - Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2High Field MR Center - Department of Biomedical Imaging and Image‐Guided Therapy, Medical University of Vienna, Vienna, Austria
In this work a Graph Neural Network for the reconstruction of undersampled k-space data is proposed. The results were evaluated and compared against a state-of-the-art method. Overall, the results suggest that this Deep Learning-based approach for reconstruction is promising.
|
||
1059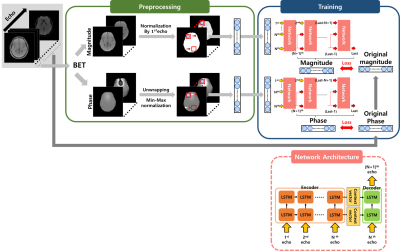 |
66 | Signal prediction in echo dimension of multi-echo gradient echo using multi-layer seq2seq model
Jisu Yun1, Seul Lee1, Muyul Park1, and Dong-Hyun Kim1
1Department of Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of The mGRE(multi-echo gradient echo) sequence has previously been used for MWF(Myelin water fraction) imaging. Such mGRE observes signal decay by obtaining multiple echo signals, but scan time increases as more echoes are obtained. To solve this trade-off, we developed a deep learning model based on a LSTM model that can reduce scan time by predicting the later echoes using only the early echoes. Looking at the in vivo results and various performance test, our network has lower RMSE and higher PSNR than NLLS(Non-linear least squares), a conventional fitting algorithm. |
||
1060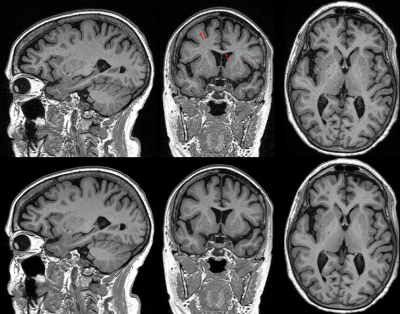 |
67 | Deep learning reconstruction enables accelerated acquisitions with consistent volumetric measurements
R. Marc Lebel1, Suryanarayanan Kaushik2, Trevor Kolupar2, Nate White3, Weidong Luo3, and Suchandrima Banerjee4
1GE Healthcare, Calgary, AB, Canada, 2GE Healthcare, Waukesha, WI, United States, 3Cortechs, San Diego, CA, United States, 4GE Healthcare, Menlo Park, CA, United States
This work evaluates brain volumetry measurements obtained from T1w images with fast (PI) and rapid (PI+CS) protocols (< 3 mins) and reconstructed with AIR Recon DL 3D, a deep learning-based reconstruction to reduce noise and ringing. Images were segmented using FDA-cleared software NeuroQuantÒ for quantitative volumetric analysis. Segmentation was successful in all cases and key brain volumes are unchanged between fast and rapid protocols, and further between conventional and DL reconstructions. This work demonstrates that highly accelerated acquisitions and advanced reconstruction methods are suitable for segmentation and volumetric studies and can improve repeatability of measurements.
|
||
1061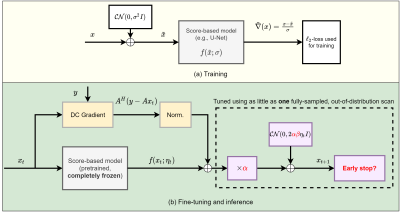 |
68 | Single-Shot Adaptation using Score-Based Models for MRI Reconstruction
Marius Arvinte1, Ajil Jalal1, Giannis Daras2, Eric Price2, Alex Dimakis1, and Jonathan I Tamir1,3,4
1Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States, 2Computer Science, The University of Texas at Austin, Austin, TX, United States, 3Oden Institute for Computational Engineering and Sciences, The University of Texas at Austin, Austin, TX, United States, 4Diagnostic Medicine, Dell Medical School, The University of Texas at Austin, Austin, TX, United States
This work deals with the problem of few-shot adaptation in data-driven MRI reconstruction, where models must efficiently adapt to new distributions. We introduce score-based models for MRI reconstruction and an algorithm for adjusting inference parameters (step size, noise level and stopping point), investigate the impact of these parameters on reconstruction performance, and demonstrate average gains of at least 2 dB in PSNR across a range of acceleration values, all while using a pretrained model that was trained for brain MRI and fine-tuned using only a single fully-sampled 2D knee scan from the fastMRI database.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.