Digital Poster
Diffusion Analysis, Tractography & Microstructure I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Computer # | ||||
|---|---|---|---|---|
0870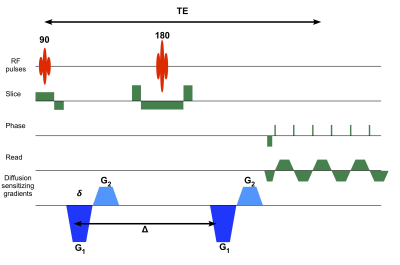 |
95 | Efficient Mapping of Diffusion Tensor Distribution in a Live Human Brain
Kulam Najmudeen Magdoom1,2,3, Alexandru V. Avram1,2,3, Dario Gasbarra4, Thomas Witzel5, Susie Y Huang5, and Peter J. Basser1,3
1Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Healt, Bethesda, MD, United States, 2The Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, United States, 3Center for Neuroscience and Regenerative Medicine, Uniformed Services University of Health Sciences, Bethesda, MD, United States, 4University of Helsinki, Helsinki, Finland, 5Athinoula A. Martinos Center for Biomedical Imaging, Boston, MA, United States
Electron microscopy of nervous tissue reveals a multitude of compartments separated by plasma membranes where the diffusion of water may be hindered. Diffusion tensor distribution (DTD) MRI assumes each voxel consists of an ensemble of diffusion tensors described by a tensor variate probability distribution function in line with the EM observed microstructure. In this study, we show in vivo results obtained in the living human brain using a new DTD framework on the Connectome scanner using 300 mT/m gradients and a novel time efficient and concomitant field free pulse sequence which allowed shorter echo time for a given b-value.
|
||
0871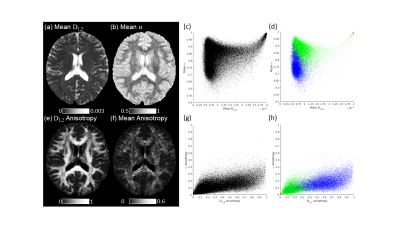 |
96 | Quasi-Diffusion Imaging: A model-based alternative to Diffusional Kurtosis Imaging
Thomas R Barrick1, Catherine A Spilling1,2, Ian R Storey1, Matt G Hall3,4, and Franklyn A Howe1
1St George's, University of London, London, United Kingdom, 2King's College London, London, United Kingdom, 3National Physical Laboratory, London, United Kingdom, 4University College London, London, United Kingdom
Quasi-Diffusion Imaging (QDI) is based on a model of diffusion dynamics that assumes diffusion is locally Gaussian within a heterogeneous tissue microstructural environment. We show here that QDI provides a compelling model-based alternative to Diffusional Kurtosis Imaging (DKI). Tensor measures determined by QDI are highly correlated with DKI, but exhibit greater parameter independence, indicating that DKI study results showing sensitivity and specificity to disease could be improved using QDI. As QDI also overcomes the limitations of DKI and can be acquired reproducibly in clinically feasible time it is a non-Gaussian diffusion imaging technique that overcomes barriers to clinical translation.
|
||
0872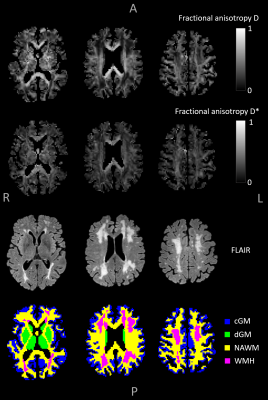 |
97 | On the anisotropy of IVIM-derived microvascular cerebral pseudo-diffusion: a physics-informed neural network approach
Paulien H.M. Voorter1,2, Jacobus F.A. Jansen1,2,3, Merel M. van der Thiel1,2, Julie Staals4,5, Robert-Jan van Oostenbrugge2,4,5, Maud van Dinther4,5, Walter H. Backes1,2,5, and Gerhard S. Drenthen1,2
1Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands, 2School for Mental Health & Neuroscience, Maastricht University, Maastricht, Netherlands, 3Department of Electrical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 4Department of Neurology, Maastricht University Medical Center, Maastricht, Netherlands, 5School for Cardiovascular Disease, Maastricht University, Maastricht, Netherlands
Acquisition of intravoxel incoherent motion (IVIM) images with diffusion sensitization in at least six directions (IVIM tensor imaging) provides the unique opportunity to non-invasively measure the anisotropy of both the parenchymal and microvascular diffusivity (D and D*). We demonstrate the feasibility of whole-brain IVIM tensor analysis by utilizing a physics-informed neural network fitting approach to achieve more accurate assessment of D, D*, and the corresponding tensors. The fractional anisotropy of D* (FA(D*)) was explored for different brain tissue regions, which revealed lower FA(D*) in cortical gray matter and higher FA(D*) in deep gray matter compared to white matter.
|
||
0873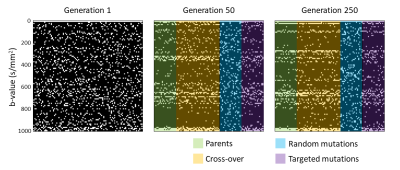 |
98 | Optimal b-value sampling for interstitial fluid estimation in cerebral IVIM imaging, a genetic algorithm approach
Gerhard S Drenthen1,2, Jacobus FA Jansen1,2, Merel M van der Thiel1,2, Paulien HM Voorter1,2, and Walter H Backes1,2
1School for Mental Health & Neuroscience, Maastricht University, Maastricht, Netherlands, 2Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands
Besides the parenchymal diffusion and microvascular pseudo-diffusion, a third diffusion component was previously found in cerebral intravoxel incoherent motion (IVIM), representing interstitial fluid. However, estimating this intermediate IVIM component can be challenging, since spectral decomposition techniques have strong dependence on the number of samples (i.e. acquired b-values) and SNR. Therefore, it is important to know which b-values are essential to be acquired. In this study, an optimal b-value sampling for estimating the intermediate component is derived using a genetic algorithm. The optimal sampling (0,20,100,270,280,370,540,650,660,710,720,790,980,990,1000 s/mm2) was shown to outperform linear and logarithmic samplings, for both simulated and in vivo data.
|
||
0874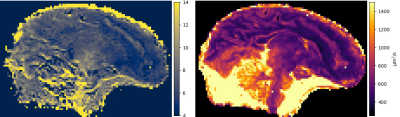 |
99 | Joint estimation of T2 and diffusion tensor from DW-SSFP using EPG
Julien Lamy1, Mathieu Santin2, and Paulo Loureiro de Sousa1
1ICube, Université de Strasbourg-CNRS, Strasbourg, France, 2Institut du Cerveau, Paris, France The DW-SSFP sequence is useful to attain strong diffusion-weighting on clinical systems while keeping a short repetition time. However, its analytical model does not account for the echo time. Using a 3D EPG model, we show that the echo time plays an important role, and that it cannot be neglected. We then provide a new method to jointly estimate T2 and the diffusion tensor on an ex-vivo fetal brain at 3 T. |
||
0875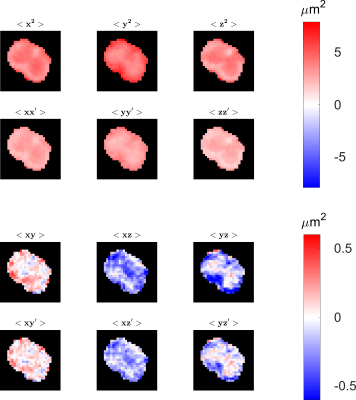 |
100 | MR measurement of the mean-position distribution
Alfredo Ordinola1 and Evren Özarslan1
1Department of Biomedical Engineering, Linköping University, Linköping, Sweden
The distribution of net displacements, commonly referred to as the ensemble average propagator, has been measured using Stejskal-Tanner experiments. Here, a recently introduced diffusion sensitization method is employed in a similar manner to obtain the distribution of mean positions. By utilizing the two schemes synergistically, low b-value measurements are employed to decouple dynamic second moments from static ones. We illustrate our findings on an excised mouse spinal cord imaged using a benchtop MRI scanner.
|
||
0876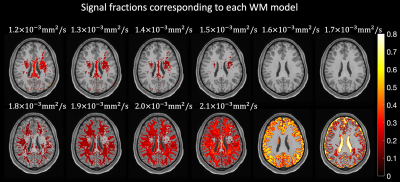 |
101 | Resolving heterogeneous crossing fibers with Adaptive modelling and Generalized Richardson Lucy spherical deconvolution (AGRL)
Alberto De Luca1,2, Alexander Leemans2, Chantal MW Tax2,3, Kurt G Schilling4, and Geert-Jan Biessels1
1Neurology Department, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht, Netherlands, 2Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 3Cardiff University Brain Research Imaging Centre (CUBRIC), Cardiff University, Cardiff, United Kingdom, 4Department of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN, United States
We evaluate the benefits of shifting from a global white matter (WM) model to an adaptive (voxel-wise) model in the Generalized Richardson Lucy (GRL) framework. Using simulations, we show that GRL with an adaptive model (AGRL) could resolve crossing fiber configurations with heterogeneous properties, whereas conventional GRL did not. In in-vivo data, AGRL simultaneously used different deconvolution models. Compared to GRL, fiber orientation distributions of AGRL showed remarkable angular differences, especially for the second and third peak. Tractography with AGRL resulted in a more extensive reconstruction of the arcuate fasciculus, suggesting adaptive modelling as a promising future direction.
|
||
0877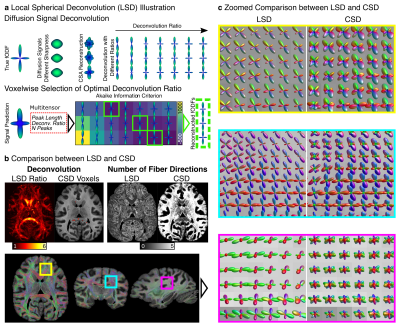 |
102 | Does One Size Fit All? Reconstructing Crossing Fibers in Diffusion MRI using Spherical Deconvolution with Local Response Functions Video Not Available
Cornelius Eichner1, Michael Paquette1, Hannah Gerbeth1, Carsten Jäger2, Christian Bock3, Guillermo Gallardo1, Torsten Möller4, Catherine Crockford5,6, Roman Wittig6, Nikolaus Weiskopf2,7, Angela D Friederici1, and Alfred Anwander1
1Department of Neuropsychology, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 3Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research, Bremerhaven, Germany, 4Kolmården Wildlife Park, Norrköping, Sweden, 5CNRS Institute of Cognitive Sciences, Lyon, France, 6Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 7Felix Bloch Institute for Solid State Physics, Faculty of Physics and Earth Sciences, Leipzig University, Leipzig, Germany
We present Local Spherical Deconvolution (LSD), a diffusion MRI deconvolution method to reconstruct crossing fiber directions in the brain. In contrast to previous approaches which assumed a single deconvolution kernel for the entire brain, LSD utilizes information theory to identify an optimal kernel in each image voxel. Using a high-resolution post-mortem chimpanzee brain, we show that fibers are reconstructed with LSD with increased precision and a reduced number of false peaks compared to conventional methods. A test-retest analysis supports the stability and accuracy of LSD.
|
||
0878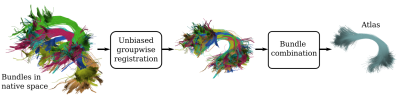 |
103 | BundleAtlasing: unbiased population-specific atlasing of bundles in streamline space
David Romero-Bascones1, Bramsh Qamar Chandio2, Shreyas Fadnavis2, Jong Sung Park2, Serge Koudoro2, Unai Ayala1, Maitane Barrenechea1, and Eleftherios Garyfallidis2
1Biomedical Engineering Department, Mondragon Unibertsitatea, Mondragón, Spain, 2Department of Intelligent Systems Engineering, Indiana University Bloomington, Bloomington, IN, United States
White matter bundle atlases play a crucial role in the segmentation of bundles and the understanding of brain connectomes. However, the construction of streamline atlases that accurately represent the underlying population anatomy is challenging. In this work, we present BundleAtlasing, a new method to compute population-specific bundle atlases in the space of streamlines. The proposed approach is based on two key aspects: an iterative groupwise unbiased bundle registration, and a pairwise bundle combination strategy. We show that our method is able to correctly generate unbiased atlases that represent the average group anatomy of a population.
|
||
0879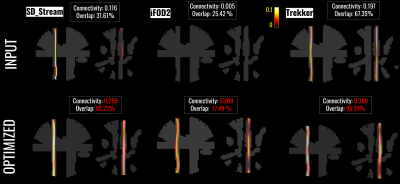 |
104 | Testing the feasibility and effectiveness of bundle-based global tractography (bundle-o-graphy) in real human brain data
Matteo Battocchio1,2, Simona Schiavi1,3, Maxime Descoteaux2, and Alessandro Daducci1
1University of Verona, Verona, Italy, 2University of Sherbrooke, Sherbrooke, QC, Canada, 3Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health (DINOGMI), University of Genoa, Genoa, Italy
We introduce a new formulation for bundle-based tractography global reconstruction (bundle-o-graphy). The idea is to move the focus from streamlines to bundle-based reconstruction thanks to a convenient reduction in the number of parameters needed to be optimized and the injection of anatomical priors. We show the potential of our approach on both synthetic and real data.
|
||
0880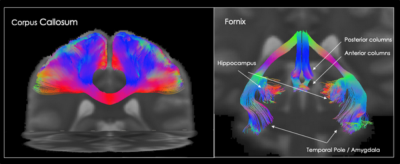 |
105 | A new framework to build high quality tractography-based DWI brain templates using COMMIT
Richard Stones1, Ahmad Beyh1,2, Rachel Barrett1, Alessandro Daducci3, and Flavio Dell'Acqua1
1Natbrainlab, King's College London, London, United Kingdom, 2University College London, London, United Kingdom, 3Universita di Verona, Verona, Italy
In this study we present a novel method for building high quality tractography-based DWI brain templates. We use streamline normalisation to obtain the correct anatomical fibre orientation information in template space and then regenerate the diffusion signal using the COMMIT framework. The template fODF field and tractography reconstructions show good anatomical agreement with well known white matter structures. This framework could provide new templates, atlases and tools for normalisation of DWI data, and simplify the creation of realistic DWI phantoms.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.