Online Gather.town Pitches
Image Reconstruction IV
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
4301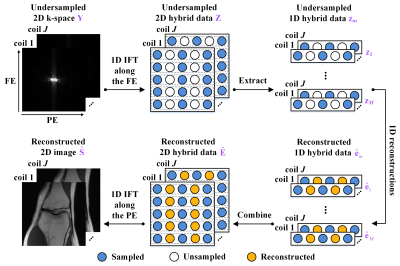 |
1 | Memory-friendly and Robust Deep Learning Architecture for Accelerated MRI
Zi Wang1, Chen Qian1, Di Guo2, Hongwei Sun3, Rushuai Li4, and Xiaobo Qu1
1Department of Electronic Science, Biomedical Intelligent Cloud R&D Center, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3United Imaging Research Institute of Intelligent Imaging, Beijing, China, 4Department of Nuclear Medicine, Nanjing First Hospital, Nanjing Medical University, Nanjing, China
Deep learning has shown astonishing performance in accelerated MRI. Most methods adopt the convolutional neural network and perform 2D convolution since many MR images or their corresponding k-space are in 2D. In this work, we try a different approach that explores the memory-friendly 1D convolution, making the deep network easier to be trained and generalized. Furthermore, a one-dimensional deep learning architecture (ODL) is proposed for MRI reconstruction. Results demonstrate that, the proposed ODL provides improved reconstructions than state-of-the-art methods and shows nice robustness to some mismatches between the training and test data.
|
||
4302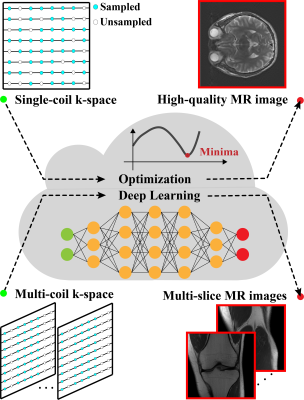 |
2 | XCloud-pFISTA: A Medical Intelligence Cloud for Accelerated MRI Reconstruction
Yirong Zhou1, Chen Qian1, Yi Guo2, Zi Wang1, Jian Wang1, Biao Qu3, Di Guo2, Yongfu You4, and Xiaobo Qu1
1Biomedical Intelligent Cloud R&D Center, Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3Department of Instrumental and Electrical Engineering, Xiamen University, Xiamen, China, 4Biomedical Intelligent Cloud R&D Center, School of Electronic Science and Engineering, China Mobile Group, Xiamen, China
Machine learning and artificial intelligence have shown remarkable performance in accelerated magnetic resonance imaging (MRI). Cloud computing technologies have great advantages in building an easily accessible platform to deploy advanced algorithms. In this work, we develop a high-performance medical intelligence cloud computing platform (XCloud-pFISTA) to reconstruct MRI images from undersampled k-space data. Two state-of-the-art approaches of the Projected Fast Iterative Soft-Thresholding Algorithm (pFISTA) family have been successfully implemented on the cloud. This work can be considered as a good example of cloud-based medical image reconstruction and may benefit the future development of integrated reconstruction and online diagnosis system.
|
||
4303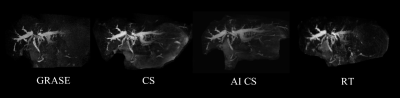 |
3 | Rapid 3D MR cholangiopancreatography in a breath-hold using deep learning constrained Compressed SENSE reconstruction
Yu Zhang1, Chunchao Xia1, Xiaoyong Zhang2, and Zhenlin Li1
1Department of Radiology, West China Hospital of Sichuan University, Chengdu, China, 2Clinical Science, Philips Healthcare, Chengdu, China
In this work, we aimed to use a deep learning based Compressed SENSE reconstruction algorithm, presented here as Artificial Intelligence Compressed-SENSE (AI-CS), to improve image quality of 3D magnetic resonance cholangiopancreatography (MRCP) in a breath hold (BH). The results demonstrated that AI-CS BH MRCP can enable improved image quality and show great visibility of small ductal structures than other previous methods.
|
||
4304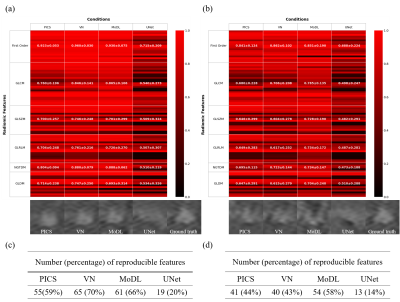 |
4 | Radiomic feature-based assessment of deep learning-based compressed sensing reconstruction
Tomoki Miyasaka1, Satoshi Funayama2, Daiki Tamada2, Hiroyuki Morisaka2, Hiroshi Onishi2, and Yasuhiko Terada1
1Graduate School of Science and Technology, University of Tsukuba, Tsukuba, Japan, 2Department of Radiology, University of Yamanashi, Chuo, Japan
Deep learning has been attracting attention as a new tool for image reconstruction. However, there is a lack of appropriate automatic evaluation metrics for reconstruction performance of small structures such as lesions, which poses a high hurdle for clinical application. Here, we explored the relationship between radiomic features of tumors and various DL reconstruction conditions, and proposed a new method based on radiomics to evaluate the reconstruction performance of DL against lesions. Based on the analysis using the concordance correlation coefficients for ground truth images, we explored several texture features that are sensitive to differences in reconstruction methods and conditions.
|
||
4305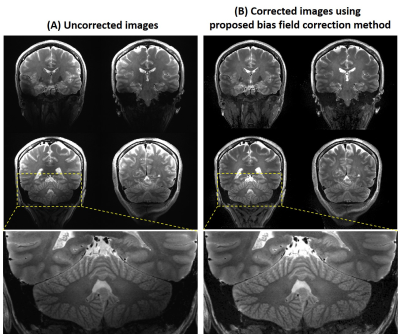 |
5 | B1- and B1+ based bias field correction for ultra-high field MRI
Yi-Cheng Hsu1, Patrick Liebig2, and Ying-Hua Chu1
1Siemens Healthineers Ltd., Shanghai, China, 2Siemens Healthcare GmbH, Erlangen, Germany
We proposed a method to approximate the magnitude of B1- and correct the bias field induced signal inhomogeneity at 7T. For 2D and 3D T2 weighted images, the signal intensity was more homogeneous and the signal void was recovered using the proposed method. This method can be implemented to correct bias field effect for other imaging methods using the estimated |B1-| and |B1+|.
|
||
4306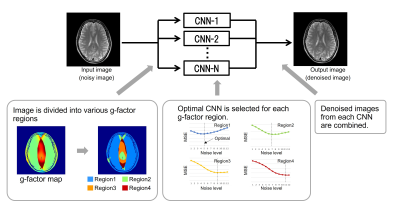 |
6 | Multi-Adaptive Convolutional Neural Network Reconstruction (MA-CNNR) for Parallel Imaging at 1.5T Brain Images
Yukio Kaneko1, Atsuro Suzuki1, Tomoki Amemiya1, Chizue Ishihara1, Yoshitaka Bito2, and Toru Shirai1
1Innovative Technology Laboratory, FUJIFILM Healthcare Corporation, Tokyo, Japan, 2Radiation Diagnostic Systems Division, FUJIFILM Healthcare Corporation, Tokyo, Japan
Recently, deep learning techniques for high-speed or high-quality imaging in MRI have been reported. However, deep learning techniques for the inhomogeneous spatial distribution of noise caused by parallel imaging have not been fully established. In this study, “Multi-Adaptive Convolutional Neural Network Reconstruction (MA-CNNR)” has been investigated. A noisy image was segmented into four regions by g-factor map, and different optimized CNNs were selected for each region. A denoised image was generated by combining the four denoised regions. The denoising effect was evaluated for 1.5T brain images, and it was confirmed that MA-CNNR can reduce the inhomogeneous noise in parallel imaging.
|
||
4307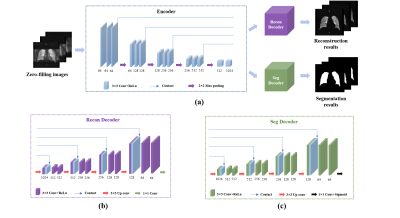 |
7 | Accelerate Pulmonary Hyperpolarized Gas MRI with Multi-Task Learning
Zimeng Li1,2, Sa Xiao2, Cheng Wang2, Chaohui Ye1,2, and Xin Zhou2
1School of Physics, Huazhong University of Science and Technology, Wuhan, China, 2Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Center for Magnetic Resonance in Wuhan, Wuhan Institute of Physics and Mathematics, Innovation Academy for Precision Measurement Science and Technology, Wuhan, China
Hyperpolarized gas MRI is a non-invasive and non-radiation imaging modality that can provide lung structure and function information. However, the problem of long imaging time limits its clinical application. Deep learning-based methods have shown great potential to accelerate MRI. In this work, we proposed a multi-task network to perform pulmonary hyperpolarized gas MRI reconstruction and lung region segmentation simultaneously, which allows sharing representations between two tasks. The results show that the proposed multi-task network has better reconstruction performance, stronger robustness and fewer model parameters than the comparison method.
|
||
4308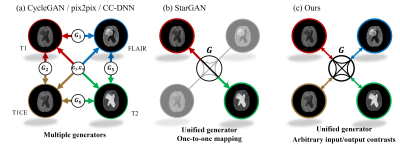 |
8 | Arbitrary Missing Contrast Generation Using Multi-Contrast Generative Network with An Encoder Network
Geonhui Son1, Yohan Jun1, Sewon Kim1, Dosik Hwang1, and Taejoon Eo*1
1Electrical and Electronic Engineering, Yonsei university, Seoul, Korea, Republic of
Multi-contrast images acquired with magnetic resonance imaging (MRI) provide abundant diagnostic information. However, the applicability of multi-contrast MRI is often limited by slow acquisition speed and high scanning cost. To overcome this issue, we propose a contrast generation method for arbitrary missing contrast images. First, StyleGAN2-based multi-contrast generator is trained to generate paired multi-contrast images. Second, pSp-based encoder network is used to predict style vectors from input images. Consequently, the imputation for arbitrary missing contrast is achieved by the process of (1) embedding one or more kinds of contrast images and (2) forward-propagating the style vector to the multi-contrast generator.
|
||
4309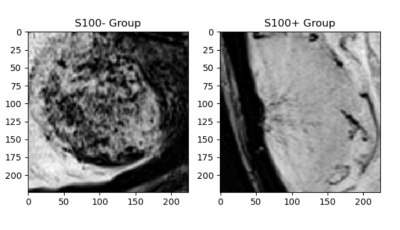 |
9 | Susceptibility Weighted MRI for Predicting Critical Developmental Regulatory S100 Proteins in Meningiomas at 3T
Sena Azamat1,2, Buse Buz-Yaluğ1, Abdullah Baş1, Alpay Ozcan3, Ayça Ersen Danyeli4,5, Kubra Tan6, Ozge Can7, Necmettin Pamir5,8, Alp Dinçer5,9, Koray Ozduman5,8, and Esin Ozturk-Isik1
1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Radiology, Basaksehir Cam and Sakura City Hospital, Istanbul, Turkey, 3Electric and Electronic Engineering Department, Bogazici University, Istanbul, Turkey, 4Department of Medical Pathology, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 5Center for Neuroradiological Applications and Reseach, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 6Health Institutes of Turkey, Istanbul, Turkey, 7Department of Medical Engineering, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 8Department of Neurosurgery, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 9Department of Radiology, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey
Meningiomas are the most common primary intracranial tumors in adults. S100 protein expression (S100+) in meningiomas is a marker of neural crest origin. Eighty-four patients with preoperative MRI were included in this IRB approved study. The whole tumor volumes were segmented from FLAIR, followed by co-registration onto SWI. Supervised machine and deep learning methods were employed to categorize meningiomas into S100+ and S100- groups based on SWI histogram values. Ensemble bagged trees resulted in an accuracy of 85.7% (sensitivity=87.0 % and specificity=84.4 %), while a Resnet 50 architecture had 70.5% accuracy (sensitivity=80%, specificity=57.1%) for predicting S100 protein expression in meningiomas.
|
||
4310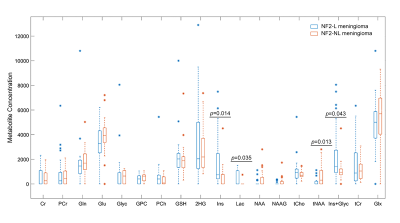 |
10 | Identification of NF2 loss in meningiomas using 1H-MRS at 3T
Banu Sacli-Bilmez1, Abdullah Baş2, Kübra Tan3, Ayça Erşen Danyeli4,5, Özge Can6, M.Necmettin Pamir5,7, Alp Dinçer5,8, Koray Özduman5,7, and Esin Ozturk-Isik1
1Institute of Biomedical Engineering, Bogazici University, İstanbul, Turkey, 2Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 3Health Institutes of Turkey, İstanbul, Turkey, 4Department of Medical Pathology, Acıbadem Mehmet Ali Aydınlar University, İstanbul, Turkey, 5Center for Neuroradiological Applications and Reseach, Acıbadem Mehmet Ali Aydınlar University, İstanbul, Turkey, 6Department of Medical Engineering, Acıbadem Mehmet Ali Aydınlar University, İstanbul, Turkey, 7Department of Neurosurgery, Acıbadem Mehmet Ali Aydınlar University, İstanbul, Turkey, 8Department of Radiology, Acıbadem Mehmet Ali Aydınlar University, İstanbul, Turkey
Loss of neurofibromatosis 2 (NF2-L) is a well-known genetic alteration of meningiomas and causes meningiomas to evolve into more aggressive and infiltrating form. This study aims to investigate single-voxel proton magnetic resonance spectroscopy (1H-MRS) correlations of NF2-L in meningiomas and to develop machine learning and deep learning models to identify NF2-L in meningiomas. NF2-L meningiomas had significantly higher Ins, Lac, and Ins+Glyc, and lower tNAA than tumors with no copy number loss. While a subspace discriminant model achieved a classification accuracy of 77.25%, a 1D-CNN model obtained a classifcation accuracy of 88.9% for identifying NF2-L meningiomas.
|
||
4311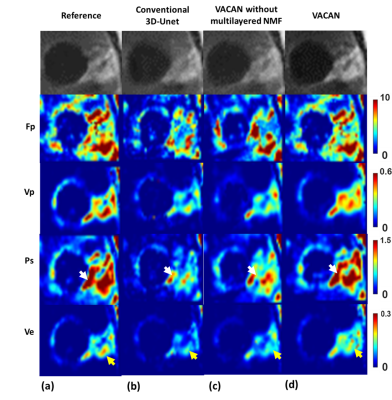 |
11 | Vascular Heterogeneity Model-based Deep Learning Reconstruction for High-Definition Dynamic Contrast Enhanced MRI
Nhan Duc Nguyen1, Joon Sik Park1, Seung Hong Choi2, Roh-Eul Yoo2, and Jaeseok Park1,3
1Department of Intelligent Precision Healthcare Convergence, Sungkyunkwan University, Suwon, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of, 3Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of
To take characterization of various vascular contrast dynamics into account, in this work we propose a novel, vascular heterogeneity model based deep learning reconstruction from highly undersampled data for high-definition whole brain DCE MRI. To this end, we introduce a new, vascular contrast dynamics (VCD) weighted deep attention neural network (VACAN) consisting of: 1) a vascular adaptive attention 3D U-Net, 2) a multilayered non-negative matrix factorization (NMF) layer, and 3) a data consistency layer. Experimental studies are performed using highly undersampled patient data to validate the effectiveness of the proposed VACAN against conventional 3D U-Net.
|
||
4312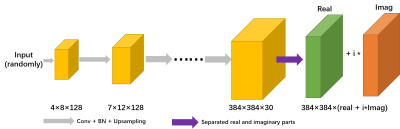 |
12 | Accelerated VCC-Wave MR Imaging Using Deep Generative Models
Congcong Liu1,2, Zhuoxu Cui1, Zhilang Qiu1, Haoxiang Li1,2, Yifan Guo1, Chentao Cao1,2, Xin Liu1,2, Hairong Zheng1,2, Dong Liang1,2,3, and Haifeng Wang1,2
1Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences, Shenzhen, China, 2Shenzhen College of Advanced Technology, University of Chinese Academy of Sciences, Shenzhen, China, 3Research Centre for Medical AI, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
CSM or kernel needs to be estimated in conventional parallel image, which is very time-consuming and the estimation process may be inaccurate. Here, wave coding model based on virtual conjugate coil using deep generative modes (WV-DGM) is proposed for the virtual conjugate coil (VCC) extended model based on wave coding. WV-DGM combined with deep DGM without training can realize advantages of wave encoding and introduce extra phase to reduce the ill-condition of the model via VCC. The results in vivo demonstrated that WV-DGM can achieve better quality compared with conventional SENSE while 10x times used to estimate CSM is reduced.
|
||
4313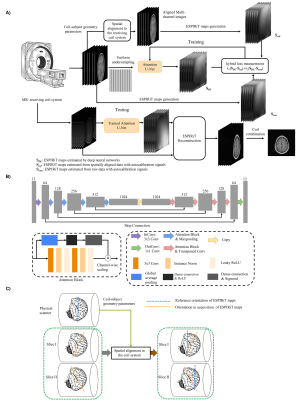 |
13 | Calibrationless Reconstruction of Uniformly Undersampled Multi-channel MR data with Deep learning Estimation of ESPIRiT Maps Video Not Available
Junhao Zhang1,2, Zheyuan Yi1,2, Yujiao Zhao1,2, Linfang Xiao1,2, Jiahao Hu1,2, Christopher Man1,2, Yujiao Zhao3, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3The University of HongKong, HongKong, China
Conventional ESPIRiT reconstruction requires accurate estimation of ESPIRiT maps from autocalibration samples or signals but acquiring such autocalibration signals takes time and may not be straightforward in some situations. This study aims to deploy deep learning to directly estimate ESPIRiT maps from uniformly undersampled multi-channel 2D MR data that contain no autocalibration signals. Results show that the estimated ESPIRiT maps could be reliably obtained and they could be used for ESPIRiT and SENSE reconstruction with high acceleration.
|
||
4314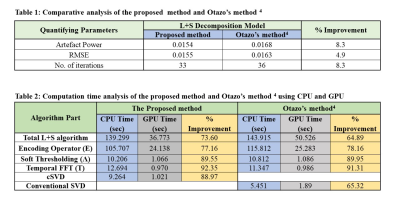 |
14 | Compressed SVD for L+S Matrix Decomposition Model to Reconstruct Undersampled Dynamic MRI
Muhammad Shafique1,2, Sohaib Ayyaz Qazi1,3, Irfan Ullah1, and Hammad Omer1
1Electrical and Computer Engineering, COMSATS University Islamabad, Islamabad, Pakistan, 2Electrical Engineering, University of the Poonch Rawalakot, Rawalakot, Pakistan, 3Service of Radiology and Faculty of Medicine, University of Geneva University Switzerland, Geneva, Swaziland
The Low rank and Sparse (L+S) matrix decomposition model has been proposed in literature to reconstruct the undersampled dynamic MRI data. The limitations of L+S method include an effective separation of the low-rank and sparse components from the acquired dynamic MRI data; also the algorithm is computationally expensive. In this paper, Compressed Singular Value Decomposition (cSVD) is employed in L+S method. The results show that the proposed method provides effective separation of the L and S components as well as considerably reduces the computation time.
|
||
4315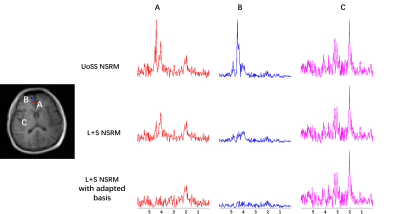 |
15 | Improved Nuisance Signal removal for 1H-MRSI Using a Low-Rank Plus Sparse Model with Learned Subspaces
Xinyu Ye1,2, Zepeng Wang2,3, and Fan Lam2,3
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 2Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, Urbana, IL, United States, 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Removing nuisance signals is an essential step for MRSI. A union-of-subspaces model that uses spatiospectral priors has achieved excellent water/lipid removal performance for 1H-MRSI, but may not be sufficient when the initial water/lipids are too strong and/or when field inhomogeneity is severe. We propose a low-rank plus sparse method for improved nuisance removal. The sparsity term is used to capture residual nuisance failed to be captured by the union-of-subspace model and the low-rank term with learned subspaces protects metabolite signals. Results from in vivo 1H-MRSI data show that the proposed method led to improved nuisance signal removal.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.