Online Gather.town Pitches
Processing & Analysis IV
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
4983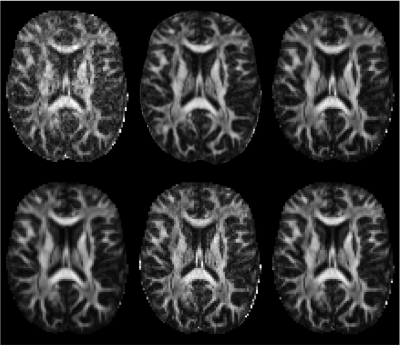 |
1 | A data-driven variability assessment of brain diffusion MRI preprocessing pipelines
Jelle Veraart1, Daan Christiaens2, Erpeng Dai3, Luke J. Edwards4, Vladimir Golkov5, Siawoosh Mohammadi6, Kurt G. Schilling7, Mohammad Hadi Aarabi8, Benjamin Ades-Aron1,9, Nagesh Adluru10, Sahar Ahmad11, Santiago Aja-Fernandez12, Andrew L. Alexander10,
Mariam Andersson13, Elixabete Ansorena14, Fakhereh Movahedian Attar4, Arnaud Attye15, Dogu Baran Aydogan16,17, Steven H. Baete1, Gianpaolo Antonio Basile18, Giulia Bertò19, Ahmad Beyh20, Alberto Cacciola18, Maxime Chamberland21, Fernando Calamante22, Jenny Chen1,
Jian Chen23, Shin Tai Chong24, Santiago Coelho1, Luis Concha25, Ricardo Coronado-Leija1, Michiel Cottaar26, Daniel Cremers5, Szabolcs David14,27, Alberto De Luca14, Flavio Dell'Acqua20, Thijs Dhollander23, Jason Druzgal28, Tim B. Dyrby13,29,
Hernández-Torres Enedino13, Oscar Esteban30, Els Fieremans1, Leon Fonville31, Björn Fricke32, Martijn Froeling33, Ian Galea34, Gabriel Girard35,36, Francesco Grussu37, Chin-Chin Heather Hsu38,39, Yung-Chin Hsu40, Khoi Huynh11, Matilde Inglese41,42,
Heide Johansen-berg26, Derek K Jones43, Kouhei Kamiya44, Claire Kelly45,46, Ahmad Raza Khan47, Ali Khan48, Yi-Chia Kung24, Alberto Lazari26, Alexander Leemans14, Laura Mancini49,50, Ivan I. Maximov51, Harri Merisaari52, Malwina Molendowska43,
Benjamin T. Newman28, Michael D. Noseworthy53, Dmitry S. Novikov1, Raquel Perez-Lopez37, Franco Pestili19, Tomasz Pieciak12, Marco Pizzolato29, Álvaro Planchuelo-Gómez12, Paul Polak54, Erika P. Raven1, Ricardo Rios-Carrillo25, Viljami Sairanen55, Simona Schiavi41,
Pohchoo Seow56, Dmitri Shastin43, Yao-Chia Shih56,57, Lucas Soustelle58, Yue Sun59, Karsten Tabelow60, Chantal MW Tax14,43, Guillaume Theaud61, Sjoerd B. Vos62, Ryckie G. Wade63, Li Wang59, Limei Wang59, Thomas Welton64,65,
Lars T. Westlye66, Stefan Winzeck67,68, Joseph Yuan-Mou Yang69,70, Pew-Thian Yap11, Yukai Zou34, Jennifer A. McNab3, Bennett A. Landman7, Nikolaus Weiskopf4,71, and Maxime Descoteaux61
1Center for Biomedical Imaging, Dept. Radiology, NYU School of Medicine, New York City, NY, United States, 2Department of Electrical Engineering (ESAT-PSI), KU Leuven, Leuven, Belgium, 3Department of Radiology, Stanford University, Stanford, CA, United States, 4Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 5Computer Vision Group, Technical University of Munich, Munich, Germany, 6Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 7Vanderbilt University Medical Center, Nashville, TN, United States, 8Department of Neuroscience and Padova Neuroscience Center, University of Padova, Padua, Italy, 9Electrical and Computer Engineering, New York University, Brooklyn, NY, United States, 10Waisman Center and Department of Radiology, UW-Madison, Madison, WI, United States, 11Department of Radiology and Biomedical Research Imaging Center (BRIC), University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 12LPI, ETSI Telecomunicacion, Universidad de Valladolid, Valladolid, Spain, 13Danish Research Centre for Magnetic Resonance, Copenhagen University Hospital Amager and Hvidovre, Copenhagen, Denmark, 14Image Sciences Institute, UMC Utrecht, Utrecht, Netherlands, 15Sydney Imaging, University of Sydney, Sydney, Australia, 16A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland, Kuopio, Finland, 17Department of Biomedical Engineering, Aalto University, Espoo, Finland, 18Brain Mapping Lab, Department of Biomedical, Dental Sciences and Morphological and Functional Images, University of Messina, Messina, Italy, 19Department of Psychology, The University of Texas at Austin, Austin, TX, United States, 20NatBrainLab, Department of Forensic and Neurodevelopmental Sciences, King's College London, London, United Kingdom, 21Donders Institute for Brain, Cognition, and Behavior, Radboud University, Nijmegen, Netherlands, 22Sydney Imaging and School of Biomedical Engineering, University of Sydney, Sydney, Australia, 23Developmental Imaging, Murdoch Children's Research Institute, Melbourne, Australia, 24Institute of Neuroscience, National Yang Ming Chiao Tung University, Tapei, Taiwan, 25Institute of Neurobiology, Universidad Nacional Autónoma de México, Querétaro, Mexico, 26Nuffield Department of Clinical Neurosciences, University of Oxford, Wellcome Centre for Integrative Neuroimaging, FMRIB, Oxford, United Kingdom, 27Department of Radiation Oncology, UMC Utrecht, Utrecht, Netherlands, 28Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, United States, 29Department of Applied Mathematics and Computer Science, Technical University of Denmark, Kongens Lyngby, Denmark, 30Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 31Department of Brain Sciences, Division of Psychiatry, Imperial College London, London, United Kingdom, 32Department of Systems Neuroscienc, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 33Department of Radiology, University medical center Utrecht, Utrecht, Netherlands, 34Department of Clinical Neurosciences, University of Southampton, Southampton, United Kingdom, 35CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 36Radiology Department, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland, 37Radiomics Group, Vall d’Hebron Institute of Oncology, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain, 38Institute of Neuroscience, National Yang Ming Chiao Tung University, Taipei, Taiwan, 39Center for Geriatrics and Gerontology, Taipei Veterans General Hospital, Tapei, Taiwan, 40AcroViz Inc., Taipei, Taiwan, 41Department of Neurosciences, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health (DiNOGMI), University of Genoa, Genoa, Italy, 42IRCCS Ospedale San Martino, Genoa, Italy, 43Cardiff University Brain Research Imaging Centre (CUBRIC), Cardiff University, Cardiff, United Kingdom, 44Department of Radiology, Toho University, Tokyo, Japan, 45Turner Institute for Brain and Mental Health, School of Psychological Sciences, Monash University, Melbourne, Australia, 46Victorian Infant Brain Studies, Murdoch Children's Research Institute, Melbourne, Australia, 47Department of Advanced Spectroscopy and Imaging, Centre of Biomedical Research, Lucknow, India, 48Department of Medical Biophysics, Western University, London, ON, Canada, 49Lysholm Department of Neuroradiology, National Hospital for Neurology and Neurosurgery UCLH NHS FT, London, United Kingdom, 50Brain Repair and Rehabilitation, University College London, London, United Kingdom, 51Western Norway University of Applied Sciences, Bergen, Norway, 52Department of Clinical Medicine, University of Turku, Turku, Finland, 53Electrical and Computer Engineering, McMaster University, Hamilton, ON, Canada, 54School of Biomedical Engineering, McMaster University, Hamilton, ON, Canada, 55BABA Center, Pediatric Research Center, Department of Clinical Neurophysiology, Children’s Hospital, University of Helsinki and Helsinki University Hospital, Helsinki, Finland, 56Diagnostic Radiology, Singapore General Hospital, Singapore, Singapore, 57Graduate Institute of Medicine, Yuan Ze University, Taoyuan City, Taiwan, 58Aix Marseille Univ, CNRS, CRMBM, Marseille, France, 59Developing Brain Computing Lab, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 60Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 61Sherbrooke Connectivity Imaging Laboratory (SCIL), Université de Sherbrooke, Sherbrooke, QC, Canada, 62Centre for Medical Image Computing, University College London, London, United Kingdom, 63Leeds Institute for Medical Research, University of Leeds, Leeds, United Kingdom, 64National Neuroscience Institute, Singapore, Singapore, 65Duke-NUS Medical School, Singapore, Singapore, 66Department of Psychology, University of Oslo, Oslo, Norway, 67Department of Computing, Imperial College London, London, United Kingdom, 68Department of Medicine, University Division of Anaesthesia, University of Cambridge, Cambridge, United Kingdom, 69Department of Neurosurgery, The Royal Children's Hospital, Melbourne, Australia, 70Neuroscience Research, Murdoch Children's Research Institute, Melbourne, Australia, 71Leipzig University, Felix Bloch Institute for Solid State Physics, Faculty of Physics and Earth Sciences, Leipzig, Germany
The preprocessing of dMRI data sets is a critical step in the experimental workflow that, in general, improves the data reliability. We provide a comprehensive survey of the preprocessing workflows for dMRI data within our research community, assessing their variability and quantifying its impact on the dMRI metric reproducibility and inter-site comparisons. We observed that the lack of a standardized preprocessing pipeline across researchers is a significant source of variability that might lower the reproducibility of studies. These preliminary results highlight the need for harmonization of the image preprocessing workflow for dMRI data to promote more robust and reproducible analyses.
|
||
4984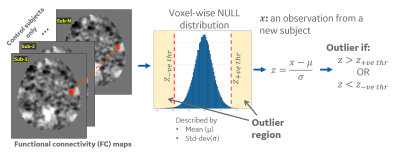 |
2 | Subject-specific multi-echo fMRI derived seed-based connectivity as a biomarker for mild traumatic brain injury Video Permission Withheld
Chitresh Bhushan1, Radhika Madhavan2, Amod Jog3, Nastaren Abad1, Brice Fernandez4, Luca Marinelli1, H. Doug Morris5, Maureen Hood5,6, J Kevin DeMarco6, Robert Y Shih5,6, Gail Kohls6, Kimbra Kenney5,6, Vincent B Ho5,6, and
Thomas K Foo1,5
1GE Research, Niskayuna, NY, United States, 2GE Healthcare, Niskayuna, NY, United States, 3GE Research, Bangalore, India, 4GE Healthcare, Versailles, France, 5Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 6Walter Reed National Military Medical Center, Bethesda, MD, United States
Mild traumatic injury (mTBI) patients exhibit acute symptoms including headaches and memory problems that sometimes persist for months, though CT/MRI scans appear normal. The purpose of this study was to identify functional biomarkers of mTBI during the 3-months following injury using seed-based functional connectivity derived from multi-echo resting state functional MRI. In this work, we use multi-band multi-echo resting state fMRI to estimate seed-based functional connectivity. We propose a novel quantitative analysis methodology of suitable for voxel-wise single-subject assessment of disrupted functional connectivity by using an outlier-analysis approach.
|
||
4985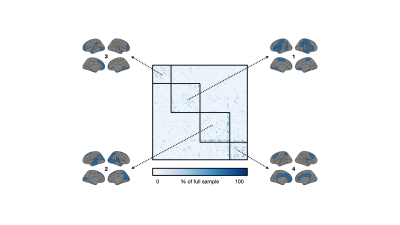 |
3 | Sample size considerations for structural covariance using T1-weighted brain imaging from the UK Biobank
William Kim1, Guocheng Jiang1,2, Nicholas Luciw1,2, and Bradley MacIntosh1,2
1Hurvitz Brain Sciences Program, Sunnybrook Research Institute, Toronto, ON, Canada, 2Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada
Anatomical T1-weighted imaging allows us to examine relationships of regional morphology across the brain through structural covariance. Here, we investigated structural covariance stability using decreasing amounts of T1-weighted imaging data from the UK Biobank. Starting from 1,753 individuals, we found that it is possible to drastically reduce the sample and still maintain adequate stability (78% agreement with ~87 individuals). We note, however, that stability was regionally variable; lateral and cortical regions were least affected by sample size while medial and subcortical regions were most affected. These findings may inform sample size considerations for MRI-based structural covariance in large population studies.
|
||
4986 |
4 | Automatically determine an optimal smoothing level in fMRI data analysis.
Xiaowei Zhuang1,2, Zhengshi Yang1, Tim Curran3, Rajesh Nandy4, Mark Lowe5, and Dietmar Cordes1,3
1Lou Ruvo Center for Brain Health, Cleveland Clinic, Las Vegas, NV, United States, 2Interdisciplinary neuroscience PhD program, University of Nevada, Las Vegas, Las Vegas, NV, United States, 3University of Colorado Boulder, Boulder, CO, United States, 4University of North Texas Health Science Center at Fort Worth, Fort Worth, TX, United States, 5Cleveland Clinic, Cleveland, OH, United States
To improve the accuracy of subject-level activation detections in noisy fMRI data, models to optimize voxel-wise smoothness levels for both isotropic Gaussian filter and spatially adaptive steerable filters are proposed. The smoothing step with currently optimized FWHM is incorporated into the optimization algorithm and solved efficiently using a sequential quadratic programming solver. Results from both simulated data and real episodic memory data indicate that a higher detection sensitivity for a fixed specificity can be achieved with the proposed method as compared to the widely used univariate general linear models with various levels of smoothness.
|
||
4987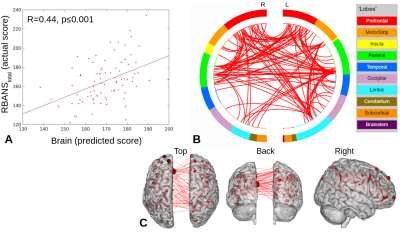 |
5 | Brain-behavior prediction using functional connectivity from older adults with mild cognitive impairment
Michelle Karker1,2, Douglas Noll1,2,3, Benjamin M. Hampstead4,5, and Scott Peltier1,2
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2Functional MRI Laboratory, University of Michigan, Ann Arbor, MI, United States, 3Radiology, University of Michigan, Ann Arbor, MI, United States, 4Mental Health Service, VA Ann Arbor Healthcare System, Ann Arbor, MI, United States, 5Research Program on Cognition and Neuromodulation Based Interventions, Psychiatry, University of Michigan, Ann Arbor, MI, United States
Partial least squares regression with feature selection (PLS-BETA) was applied to task-based and resting-state connectivity data. Leveraging a MCI-relevant object-location or face-name task illuminates relationships with measures of total cognition (RBANStotal) and memory (RBANSdelayed). This provides support for the use of clinically-relevant tasks in cases where a “driven” connectivity network may elucidate pathological changes.
|
||
4988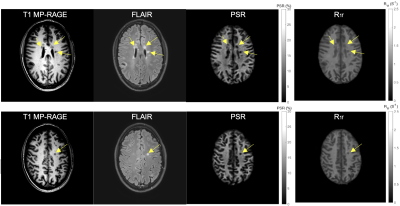 |
6 | Test-retest Repeatability of SIR-QMT Using Compressed SENSE
Ping Wang1, Nicholas J. Sisco1, Aimee Borazanci2, and Richard D. Dortch1
1Translational Neuroscience, Barrow Neurological Institute, Phoenix, AZ, United States, 2Neurology, Barrow Neurological Institute, Phoenix, AZ, United States
Quantitative magnetization transfer (QMT) imaging using the selective inversion recovery (SIR) approach has been successful in estimating fundamental tissue parameters, such as the PSR (macromolecular-to-free proton pool size ratio) and the R1f (relaxation rate of the free pool). Although SIR allows one to perform the QMT experiment using conventional inversion recovery sequences, it is hampered by long scan times. Previously we have shown that Compressed SENSE (CS-SENSE) accelerates whole-brain SIR-QMT imaging, allowing whole-brain scanning within clinically practical scan times. In this study, we systematically investigate the effect of this acceleration on test-retest repeatability for PSR and R1f.
|
||
4989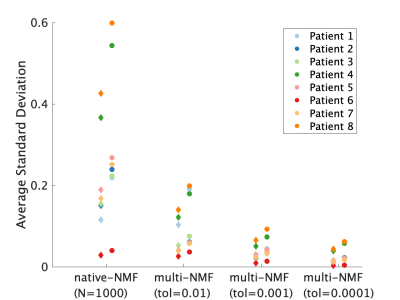 |
7 | Increased repeatability in blind source separation analysis of dynamic contrast enhanced MRI Video Permission Withheld
Dipal Patel1, Alexandru Badalan1, Zaki Ahmed1,2, and Ives R. Levesque1,3
1Medical Physics Unit, McGill University, Montreal, QC, Canada, 2Mayo Clinic, Rochester, MN, United States, 3Research Institute of the McGill University Health Centre, Montreal, QC, Canada
Blind source separation can be used to linearly decompose DCE-MRI time-course data into a sparse set of time courses, or sources, and maps of coefficients, or weights, to describe the entire 4D dataset. This type of analysis generates in realistic time-courses for the wash-in and wash-out of the contrast agent, and maps of the distribution of these dynamics. In turn, these decompositions may hold diagnostic value. Random initialization typical of such algorithms makes the output unstable. This work sought design an approach to blind source separation analysis of DCE-MRI with lower variability and independent of NMF initialization. |
||
4990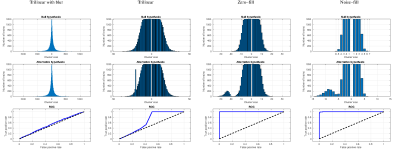 |
8 | Noise-Fill Interpolation Improves Statistical Power of Lesion Detection in Voxel-Wise Analyses
Roman Fleysher1, Lazar Fleysher2, Namhee Kim3, Michael L Lipton1, and Craig A Branch1
1Gruss Magnetic Resonance Research Center, Department of Radiology, Albert Einstein Colledge of Medicine, Bronx, NY, United States, 2Biomedical Engineering and Imaging Institute, Department of Radiology, Mount Sinai Medical Center, New York, NY, United States, 3Department of Neurological Sciences, Rush Medical College, Chicago, IL, United States
Image interpolation is inextricable in many image analysis steps including registration of low resolution images to a standard high resolution template. Interpolated images are often smoothed and their voxel intensities are not statistically independent which severely complicates subsequent statistical analysis at the group level. Difficult to account correlations lead to higher than expected false-positive rates. We propose a new, noise-fill, image interpolation method which avoids both spatial blurring and loss of statistical independence of the voxel intensities. We show that noise-fill interpolation improves sensitivity of lesion detection in simulated patient-specific voxel-wise cluster analyses compared to other typically used interpolation methods.
|
||
4991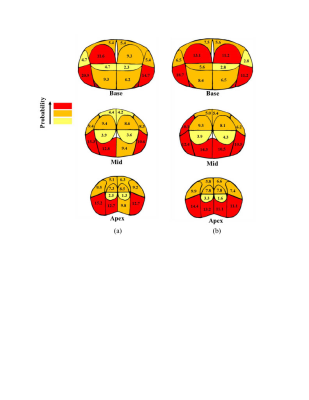 |
9 | Localization of Prostate Cancer at 3-T Multiparametric Magnetic Resonance Imaging using Prostate Sector Map
Fatemeh Zabihollahy1, Steven S. Raman1, Pornphan Wibulpolprasert2, Robert Reiter3, Holden Wu1, and Kyung Hyun Sung1
1Radiology, University of California, Los Angeles, Los Angeles, CA, United States, 22Department of Diagnostic and Therapeutic Radiology, Ramathibodi Hospital, Bangkok, Thailand, 3Urology, University of California, Los Angeles, Los Angeles, CA, United States
Multiparametric magnetic resonance imaging (mpMRI) has a significant impact on prostate cancer (PCa) diagnosis. However, it is important to realize its accuracy for PCa detection. In this study, we compare mpMRI with whole-mount histopathology (WMHP) images as a reference to discover the limitation of mpMRI for PCa lesion localization. The results are presented as a spatial probability map, corresponding to the prostate sector map used in Prostate Imaging Reporting and Data System version 2.1 (PI-RADSv2.1), to highlight the regions on the prostate glands that require further attention from clinicians for a more accurate diagnosis of PCa and its treatment planning.
|
||
4992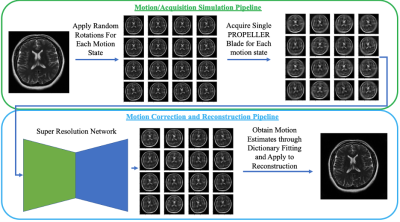 |
10 | Super Resolution Enhanced PROPELLER for Retrospective Motion Correction
Brett Levac1 and Jonathan I Tamir1,2,3
1Electrical and Computer Engineering, University of Texas, Austin, TX, United States, 2Oden Institute for Computational Engineering and Sciences, University of Texas, Austin, TX, United States, 3Department of Diagnostic Medicine, University of Texas, Austin, TX, United States
PROPELLER based acquisitions have the unique ability to give low resolution images for each echo train acquired and are often used for motion correction. However, motion correction with PROPELLER can often be hindered due to the low resolution nature of each shot. We propose a technique which leverages recent advancements in super resolution neural networks to enhance low resolution PROPELLER shots for better inter-shot motion estimation.
|
||
4993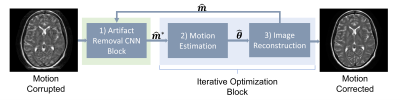 |
11 | Assessing the Role of Deep Learning in Joint Motion and Image Estimation
Brian Nghiem1,2, Zhe Wu1, Melissa Haskell3, Lars Kasper1, and Kamil Uludag1,2
1BRAIN-To Lab, University Health Network, Toronto, ON, Canada, 2Medical Biophysics, University of Toronto, Toronto, ON, Canada, 3Electrical Engineering and Computer Science, University of Michigan, Ann Arbour, MI, United States
We investigated the performance of CNN-assisted joint estimation in two cases of severe motion corruption in a 2D slice of T2w FSE MRI. We showed that the inclusion of the CNN can help speed up convergence of the joint estimation algorithm, corroborating previous findings. We also showed one case in which joint estimation failed to converge to the correct image and motion parameters, with and without the CNN. A more exhaustive study is required to confirm whether deep learning can help joint estimation salvage otherwise unsalvageable corrupted data.
|
||
4994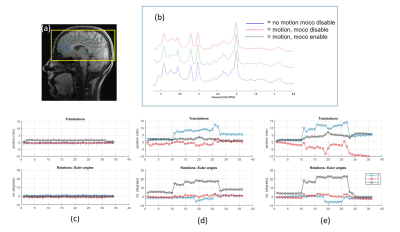 |
12 | Prospective Motion Correction for MR Spectroscopy in the Human Brain Using Multi-Slice Spiral Navigator
Nutandev Bikkamane Jayadev1, Pierre-Gilles Henry1, and Dinesh Deelchand1
1University of Minnesota, MINNEAPOLIS, MN, United States
MR spectroscopy is prone to motion artifacts due to long scan times. Existing motion correction techniques require additional hardware or are inherently slow. We demonstrate a fast and accurate prospective motion correction method for MRS using spiral navigators. A multi-slice to volume registration approach accelerates the motion parameter determination and achieves artifact free spectra at intended volume of interest.
|
||
4995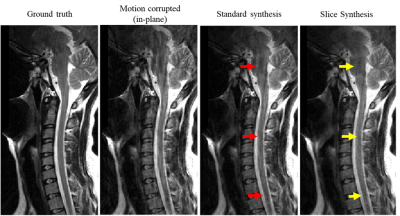 |
13 | COCOA Synthesis Using Adjacent Slices for Improved Motion Artifact Correction
Sampada Bhave1, Jennifer Wagner1, and Hassan Haji-valizadeh1
1Canon Medical Research USA, Mayfield Village, OH, United States
A modified COCOA framework was introduced to suppress motion artifacts. The data convolution operation in COCOA was extended to include k-space neighbors in the slice dimension along with those in the slice. Regularization in the data convolution operation was used as an alternative to the data combination in COCOA. The proposed technique provided high motion artifact suppression as compared to the standard COCOA synthesis.
|
||
4996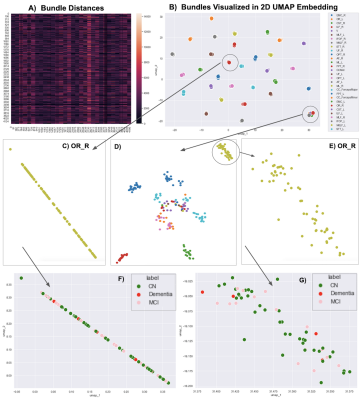 |
14 | Visualizing 4,230 White Matter Tracts at Once
Bramsh Qamar Chandio1,2, Tamoghna Chattopadhyay3, Conor Owens-Walton3, Julio E. Villalon Reina3, Leila Nabulsi3, Sophia I. Thomopoulos3, Javier Guaje4, Eleftherios Garyfallidis4, and Paul M. Thompson3
1Department of Intelligent SystemsEngineering, School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, IN, United States, 2Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Marina Del Rey, CA, United States, 3Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Marina del Rey, CA, United States, 4Department of Intelligent Systems Engineering, School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, IN, United States
We propose a dimensionality reduction method, based on the bundle-based minimum distance metric and the UMAP technique, to disentangle and visualize clusters in whole-brain tractography. In multishell diffusion MRI data from 141 elderly subjects, 30 tracts were extracted per subject using auto-calibrated RecoBundles in DIPY. A (141x30)x(141x30) bundle distance matrix was calculated and fed into UMAP. Embedding space maps showed that the same bundles were consistently mapped across subjects, making it easier to identify outliers and define clusters for population-based statistical analysis.
|
||
4997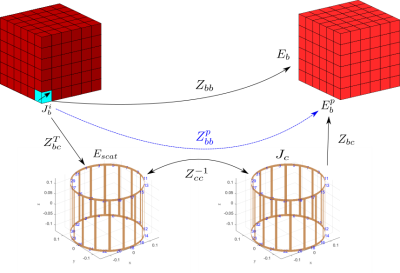 |
15 | Fast full-wave patient-specific field simulations in seconds with MARIE 2.0 Video Permission Withheld
Georgy Guryev1, Eugene Milshteyn2,3, Ilias I Giannakopoulos4,5, Elfar Adalsteinsson1,6,7, Lawrence L. Wald2,3,7, and Jacob K White1
1Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Harvard Medical School, Boston, MA, United States, 4Center for Advanced Imaging Innovation and Research (CAI2R), Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 5The Bernard and Irene Schwartz Center for Biomedical Imaging (CBI), Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 6Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 7Harvard-MIT Division of Health Sciences Technology, Cambridge, MA, United States
It was recently demonstrated that the combination of fast low-resolution tissue mapping and fast voxel-based field simulation can be used to perform a patient-specific MR safety check in minutes[4,5].However,field simulation required several of those minutes,making it too slow to perform the dozens of simulations that would be needed for patient-specific optimization.In this abstract we describe a compressed-perturbation-matrix technique that nearly eliminates the computational cost of including complex coils (or coils+shields) in voxel-based field simulation of tissue,thereby reducing simulation time from minutes to seconds.The approach is demonstrated on a wide variety of head+coil and head+coil+shield configurations,using the latest implementation of MARIE2.0.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.