Online Gather.town Pitches
Machine Learning & Artificial Intelligence III
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
 |
4344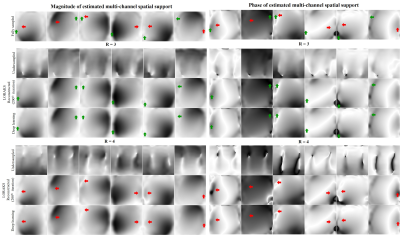 |
1 | Fast and Calibrationless Low-Rank Reconstruction through Deep Learning Estimation of Multi-Channel Spatial Support
Zheyuan Yi1,2,3, Yujiao Zhao1,2, Linfang Xiao1,2, Yilong Liu1,2, Christopher Man1,2, Jiahao Hu1,2,3, Vick Lau1,2, Alex Leong1,2, Fei Chen3, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China In traditional parallel imaging, calibration data need to be acquired, prolonging data acquisition time or/and sometimes increasing the susceptibility to motion. Low-rank parallel imaging has emerged as a calibrationless alternative that formulates reconstruction as a structured low-rank matrix completion problem while incurring a cumbersome iterative reconstruction process. This study achieves a fast and calibrationless low-rank reconstruction by estimating high-quality multi-channel spatial support directly from undersampled data via deep learning. It offers a general and effective strategy to advance low-rank parallel imaging by making calibrationless reconstruction more efficient and robust in practice. |
|
4345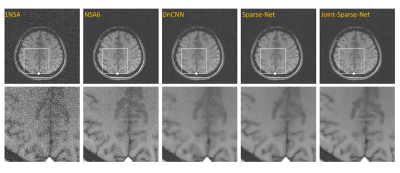 |
2 | A deep learning network for low-field MRI denoising using group sparsity information and a Noise2Noise method
Yuan Lian1, Xinyu Ye1, Hai Luo2, Ziyue Wu2, and Hua Guo1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 2Marvel Stone Healthcare Co., Ltd., Wuxi, China
Owing to hardware advancements, interest in low-field MRI system has increased recently. However, the imaging quality of low-field MRI is limited due to intrinsic low signal to noise ratio (SNR). Here we propose a deep-learning model to jointly denoise multi-contrast images using Noise2Noise training strategy. Our method can promote the SNRs of multi-contrast low-field images, and experiments show the effectiveness of the proposed strategy.
|
||
4346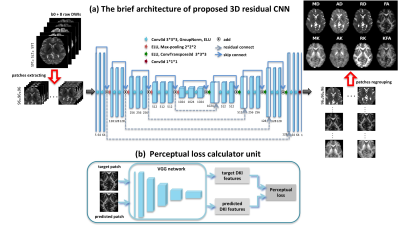 |
3 | High-quality Reconstruction of Volumetric Diffusion Kurtosis Metrics via Residual Learning Network and Perceptual Loss
Min Feng1, Qiqi Tong2, Yingying Li1, Bo Dong1, JIanhui Zhong1,3, and Hongjian He1
1Center for Brain Imaging Science and Technology, College of Biomedical Engineering and Instrumental Science, Zhejiang University, HANGZHOU, China, 2Research Center for Healthcare Data Science, Zhejiang Lab, HANGZHOU, China, 3Department of Imaging Sciences, University of Rochester, Rochester, NY, United States
This study proposed a novel 3D residual network to learn end-to-end reconstruction from as few as eight DWIs to volumetric DKI parameters. The weighted loss function combining perceptual loss is utilized, which helps the network capture in-depth feature of DKI parameters. The results show that our method achieves superior performance over state-of-the-art methods for providing accurate DKI parameters as well as preserves rich textural details and improves the visual quality of reconstructions.
|
||
4347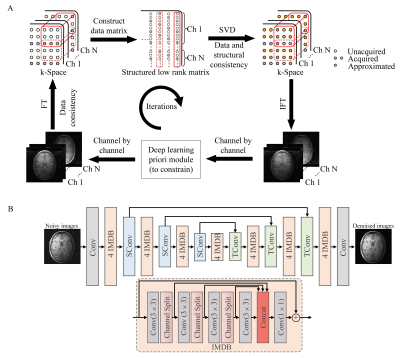 |
4 | Low-rank Parallel Imaging Reconstruction Imbedded with a Deep Learning Prior Module
Linfang Xiao1,2, Yilong Liu1,2, Zheyuan Yi1,2, Yujiao Zhao1,2, Alex T.L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China
Recently, deep learning methods have shown superior performance on image reconstruction and noise suppression by implicitly yet effectively learning prior information. However, end-to-end deep learning methods face the challenge of potential numerical instabilities and require complex application specific training. By taking advantage of the multichannel spatial encoding (as exploited by conventional parallel imaging reconstruction) and prior information (exploited by deep learning methods), we propose to embed a deep learning module into the iterative low-rank matrix completion based image reconstruction. Such strategy significantly suppresses the noise amplification and accelerates iteration convergence without image blurring.
|
||
4348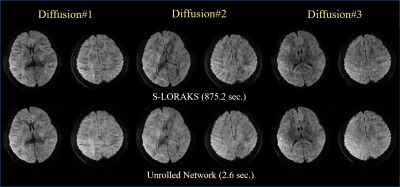 |
5 | Rapid reconstruction of Blip up-down circular EPI (BUDA-cEPI) for distortion-free dMRI using an Unrolled Network with U-Net as Priors Video Permission Withheld
Uten Yarach1, Itthi Chatnuntawech2, Congyu Liao3, Surat Teerapittayanon2, Siddharth Srinivasan Iyer4,5, Tae Hyung Kim6,7, Jaejin Cho6,7, Berkin Bilgic6,7, Yuxin Hu8, Brian Hargreaves3,8,9, and Kawin Setsompop3,8
1Department of Radiologic Technology, Faculty of Associated Medical Science, Chiang Mai University, Chiang Mai, Thailand, 2National Nanotechnology Center (NANOTEC), National Science and Technology Development Agency (NSTDA), Pathum Thani, Thailand, 3Department of Radiology, Stanford University, Stanford, CA, United States, 4Department of Radiology, Stanford University, Stanford, Stanford, CA, United States, 5Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 6Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 7Department of Radiology, Harvard Medical School, Boston, MA, United States, 8Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 9Department of Bioengineering, Stanford University, Stanford, CA, United States
Blip-up and Blip-down EPI (BUDA) is a rapid, distortion-free imaging method for diffusion-imaging and quantitative-imaging. Recently, we developed BUDA-circular-EPI (BUDA-cEPI) to shorten the readout-train and reduce T2* blurring for high-resolution applications. In this work, we further improve encoding efficiency of BUDA-cEPI by leveraging partial-Fourier in both phase-encode and readout directions, where complimentary conjugate k-space information from the blip-up and blip-down EPI-shots and S-LORAKS constraint are used to effectively fill-out missing k-space. While effective, S-LORAKS is computationally expensive. To enable clinical deployment, we also proposed a machine-learning reconstruction derived from RUN-UP (unrolled K-I network) that accelerates reconstruction by >300x.
|
||
4349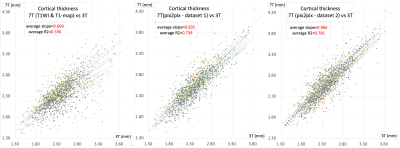 |
6 | A Deep Learning Approach to Improve 7T MRI Anatomical Image Quality Deterioration Due mainly to B1+ Inhomogeneity
Thai Akasaka1, Koji Fujimoto2, Yasutaka Fushimi3, Dinh Ha Duy Thuy1, Atsushi Shima1, Nobukatsu Sawamoto4, Yuji Nakamoto3, Tadashi Isa1, and Tomohisa Okada1
1Human Brain Research Center, Kyoto University, Kyoto, Japan, 2Department of Real World Data Research and Development, Kyoto University, Kyoto, Japan, 3Diagnostic Imaging and Nuclear Medicine, Kyoto University, Kyoto, Japan, 4Department of Human Health Sciences, Kyoto University, Kyoto, Japan
The effect of transmit field (B1+) inhomogeneity at 7T remains even after correction using a B1+-map. By a deep learning approach using pix2pix, a neural network was trained to generate 3T-like anatomical images from 7T MP2RAGE images (dataset 1: T1WI and T1-map after B1+ correction and dataset 2: Inversion time [INV]1, INV2 and B1+-map). When the HCP anatomical pipeline was applied and compared, low regressions of original 7T data to 3T data were largely improved by using generated images by pix2pix, especially for dataset 2.
|
||
4350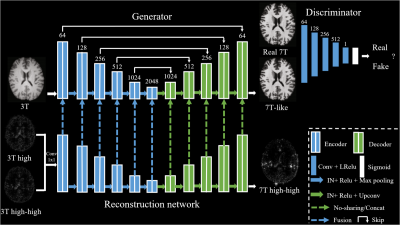 |
7 | 7T MRI prediction from 3T MRI via a high frequency generative adversarial network
Yuxiang Dai1, Wei Tang1, Ying-Hua Chu2, Chengyan Wang3, and He Wang1,3
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Siemens Healthineers Ltd., Shanghai, China, 3Human Phenome Institute, Fudan University, Shanghai, China
Existing methods often fail to capture sufficient anatomical details which lead to unsatisfactory 7T MRI predictions, especially for 3D prediction. We proposed a 3D prediction model which introduces high frequency information learned from 7T images into generative adversarial network. Specifically, the prediction model can effectively produce 7T-like images with sharper edges, better contrast and higher SNR than 3T images.
|
||
4351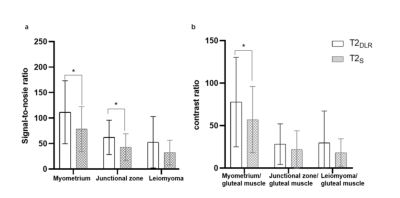 |
8 | Deep learning-accelerated T2-weighted imaging of the female pelvis: reduced acquisition times and improved image quality Video Not Available
Jing Ren1, Yonglan He1, Chong Liu1, Shifei Liu 1, Jinxia Zhu2, Marcel Dominik Nickel3, Zhengyu Jin1, and Huadan Xue1
1Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China, 2MR collaboration, Siemens Healthineers Ltd., Beijing, China, 3MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany Novel deep learning (DL) reconstruction methods may accelerate female pelvis MRI protocols keeping high image quality. The value of a novel DL reconstruction of T2-weighted (T2DLR) turbo spin-echo (TSE) sequences for female pelvis MRI in three orthogonal planes was evaluated. We evaluated examination times, image quality, and lesion conspicuity of benign uterine disease. The T2DLR quantitative parameters remained similar or were significantly improved compared with that of standard T2 TSE (T2S), allowing for a 62.7% reduction in acquisition times. Applying this novel T2DLR sequence achieved better image quality and shorter acquisition time than T2S. |
||
4352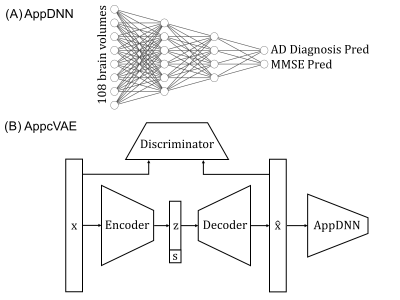 |
9 | Application-specific structural brain MRI harmonization Video Not Available
Lijun An1,2,3, Pansheng Chen1,2,3, Jianzhong Chen1,2,3, Christopher Chen4, Juan Helen Zhou2, and B.T. Thomas Yeo1,2,3,5,6
1Department of Electrical and Computer Engineering, National University of Singapore, Singapore, Singapore, 2Centre for Sleep and Cognition (CSC) & Centre for Translational Magnetic Resonance Research (TMR), National University of Singapore, Singapore, Singapore, 3N.1 Institute for Health & Institute for Digital Medicine (WisDM), National University of Singapore, Singapore, Singapore, 4Department of Pharmacology, National University of Singapore, Singapore, Singapore, 5NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore, Singapore, 6Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States
We propose a flexible application-specific harmonization framework utilizing downstream application performance to regularize the harmonization procedure. Our approach can be integrated with various deep learning models. Here, we apply our approach to the recently proposed conditional variational autoencoder (cVAE) harmonization model. Three datasets (ADNI, N=1735; AIBL, N=495; MACC, N=557) collected from three different continents were used for evaluation. Our results suggest our approach (AppcVAE) compares favorably with ComBat (named for “combating batch effects when combining batches”) and cVAE for improving downstream application performance.
|
||
4353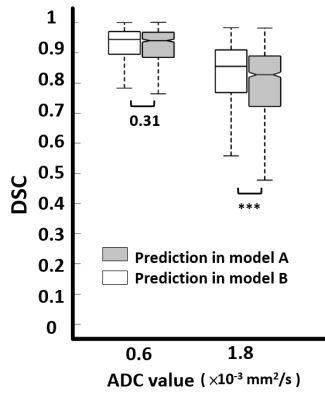 |
10 | Better Inter-observer agreement for Stroke Segmentation on DWI in Deep Learning Models than Human Experts Video Not Available
Shao Chieh Lin1,2, Chun-Jung Juan2,3,4, Ya-Hui Li2, Ming-Ting Tsai2, Chang-Hsien Liu2, Hsu-Hsia Peng5, Teng-Yi Huang6, Yi-Jui Liu7, and Chia-Ching Chang2,8
1Ph.D. program in Electrical and Communication Engineering, Feng Chia University, Taichung, Taiwan, 2Department of Medical Imaging, China Medical University Hsinchu Hospital, Hsinchu, Taiwan, 3Department of Radiology, School of Medicine, College of Medicine, China Medical University, Taichung, Taiwan, 4Department of Computer Science and Information Engineering, National Taiwan University, Taipei, Taiwan, 5Department of Biomedical Engineering and Environmental Sciences, National Tsing Hua University, Hsinchu, Taiwan, 6Department of Electrical Engineering, National Taiwan University of Science and Technology, Taipei, Taiwan, 7Department of Automatic Control Engineering, Feng Chia University, Taichung, Taiwan, 8Department of Management Science, National Yang Ming Chiao Tung University, Hsinchu, Taiwan
Inter-observer agreement is commonly used to evaluate the consistency of clinical diagnosis for two or more doctors. However, it is seldom to use to evaluate the consistency of clinical diagnosis for two or more deep learning models. In this study, four deep learning models for segmentation of stroke lesion were trained using GTs defined by two neuroradiologists with two ADC thresholds. We found the addition of an ADC threshold (0.6 × 10-3 mm2/s) helps eliminate inter-observer variation and achieve best segmentation performance. The inter-observer in two deep learning models shows the more consistent degree compared with inter-observer in two neuroradiologists.
|
||
4354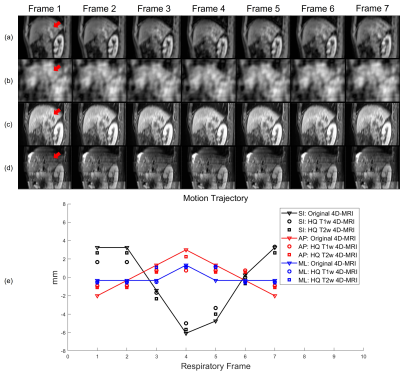 |
11 | Real-time High-quality Multi-parametric 4D-MRI Using Deep Learning-based Motion Estimation from Ultra-undersampled Radial K-space
Haonan Xiao1, Yat Lam Wong1, Wen Li1, Chenyang Liu1, Shaohua Zhi1, Weiwei Liu2, Weihu Wang2, Yibao Zhang2, Hao Wu2, Ho-Fun Victor Lee3, Lai-Yin Andy Cheung4, Hing-Chiu Charles Chang5, Tian Li1, and Jing Cai1
1Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, China, 2Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Radiation Oncology, Beijing Cancer Hospital & Institute, Peking University Cancer Hospital & Institute, Beijing, China, 3Department of Clinical Oncology, The University of Hong Kong, Hong Kong, China, 4Department of Clinical Oncology, Queen Mary Hospital, Hong Kong, China, 5Department of Radiology, The University of Hong Kong, Hong Kong, China
We have developed and validated a deep learning-based real-time high-quality (HQ) multi-parametric (Mp) 4D-MRI technique. A dual-supervised downsampling-invariant deformable registration (D3R) model was trained on retrospectively downsampled 4D-MRI with 100 radial spokes in the k-space. The deformations obtained from the downsampled 4D-MRI were applied to 3D-MRI to reconstruct HQ Mp 4D-MRI. The D3R model provides accurate and stable registration performance at up to 500 times downsampling, and the HQ Mp 4D-MRI shows significantly improved quality with sub-voxel level motion accuracy. This technique provides HQ Mp 4D-MRI within 500 ms and holds great potential in online tumor tracking in MR-guided radiotherapy.
|
||
4355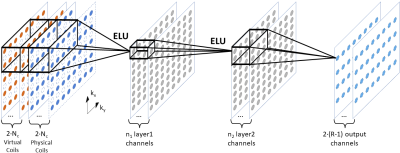 |
12 | Advancing RAKI Parallel Imaging Reconstruction with Virtual Conjugate Coil and Enhanced Non-Linearity
Christopher Man1,2, Zheyuan Yi1,2, Vick Lau1,2, Jiahao Hu1,2, Yujiao Zhao1,2, Linfang Xiao1,2, Alex T.L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China
RAKI is recently proposed as a deep learning version of GRAPPA, which trains on auto-calibration signal (ACS) to estimate the missing k-space data. However, RAKI requires a larger amount of ACS for training and reconstruction due to its multiple convolutions which resulting in lower effective acceleration. In this study, we propose to incorporate the virtual conjugate coil and enhanced non-linearity into the RAKI framework to improve the noise resilience and artifact removal at high effective acceleration. The results demonstrate that such strategy is effective and robust at high effective acceleration and in presence of pathological anomaly.
|
||
4356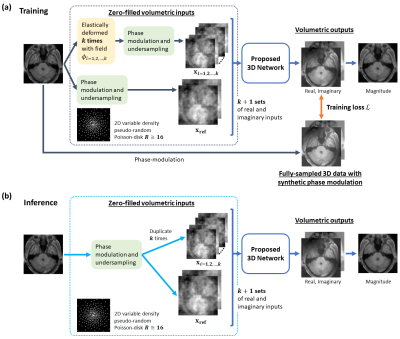 |
13 | Accelerate MR imaging by anatomy-driven deep learning image reconstruction
Vick Lau1,2, Christopher Man1,2, Yilong Liu1,2, Alex T. L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China
Supervised deep learning (DL) methods for MRI reconstruction is promising due to their improved reconstruction quality compared with traditional approaches. However, all current DL methods do not utilise anatomical features, a potentially useful prior, for regularising the network. This preliminary work presents a 3D CNN-based training framework that attempts to incorporate learning of anatomy prior to enhance model’s generalisation and its stability to perturbation. Preliminary results on single-channel HCP, unseen pathological HCP and IXI volumetric data (effective R=16) suggest its potential capability for achieving high acceleration while being robust against unseen anomalous data and data acquired from different MRI systems.
|
||
4357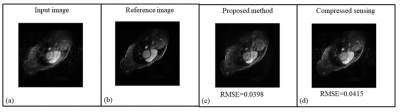 |
14 | A Hybrid Dual Domain Deep Learning Framework for Cardiac MR Image Reconstruction
Rameesha Khawaja1, Amna Ammar1, Madiha Arshad1, Faisal Najeeb1, and Hammad Omer1
1MIPRG Research Group, ECE Department, Comsats University, Islamabad, Pakistan
Reconstruction of cine Cardiac MRI (CMRI) is an active research area with room for improvement in motion detection (particularly irregular cardiac motion) and modeling in order to significantly enhance the quality of reconstructed images. Moreover, the reduction of scan time and image reconstruction time of cine CMRI is also a key aspect of today’s clinical requirement. We propose a dual domain cascade of neural networks intercalated with multi-coil data consistency layers for the reconstruction of cardiac MR images from Variable Density under-sampled data. The results show successful reconstruction results of our proposed method when compared with conventional compressed sensing reconstruction.
|
||
4358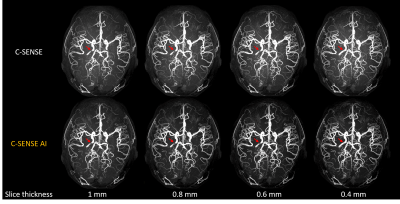 |
15 | Ultra-thin slice Time-of-Flight MR angiography for brain with a deep learning constrained Compressed SENSE reconstruction
Jihun Kwon1, Takashige Yoshida2, Masami Yoneyama1, Johannes M Peeters3, and Marc Van Cauteren3
1Philips Japan, Tokyo, Japan, 2Tokyo Metropolitan Police Hospital, Tokyo, Japan, 3Philips Healthcare, Best, Netherlands
Time-of-flight MR angiography (TOF-MRA) is a non-contrast-enhanced imaging technique widely used to visualize intracranial vasculature. In this study, we investigated the use of ultra-thin slice (up to 0.4 mm) to improve the delineation of the cerebral arteries in TOF-MRA. To reduce the noise while preserving the image quality, Compressed SENSE AI (CS-AI) reconstruction was used. Our results showed that the improved noise reduction by CS-AI enabled better visualization of vessels, especially on the thinner slices compared to conventional Compressed-SENSE. The usefulness of CS-AI was also demonstrated in clinical cases with moyamoya disease and suspected aneurysm patients.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.