Online Gather.town Pitches
Machine Learning/Artificial Intelligence I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
3187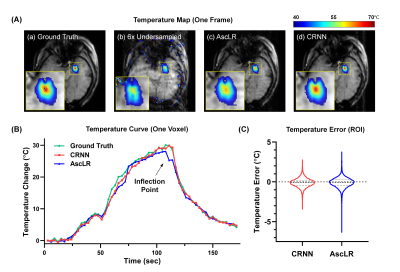 |
1 | Real-time Reconstruction for Accelerated MR Thermometry Using CRNN in MRgLITT Treatment
Ziyi Pan1, Jieying Zhang1, Kai Zhang2, Hao Sun3, Meng Han3, Yawei Kuang3, Jiaqi Dou1, Wenbo Liu3, and Hua Guo1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 2Beijing Tiantan Hospital, Capital Medical University, Beijing, China, 3Sinovation Medical, Beijing, China
MRI-guided laser-interstitial thermal therapy (MRgLITT) is a minimally invasive therapeutic method in neurosurgery. Accelerating the data acquisition of thermometry for MRgLITT treatment is crucial in achieving high temporal-spatial resolution and large volume coverage. In this work, we suggest using the convolutional recurrent neural network (CRNN) to achieve real-time reconstruction for accelerated temperature mapping, because CRNN is capable of utilizing the temporal correlations of dynamic data to resolve aliasing artifacts. Results demonstrate that 6-fold acceleration can be achieved in MRgLITT treatment using CRNN with clinically acceptable reconstruction time and temperature measurement errors.
|
||
3188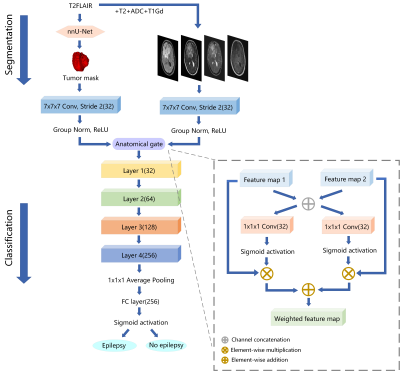 |
2 | Deep Learning Based Automated Diagnosis of Epilepsy in Patients with WHO II-IV Grade Cerebral Gliomas from Multiparametric MRI
Hongxi Yang1, Ankang Gao2, Yida Wang1, Xu Yan3, Jingliang Cheng2, Jie Bai2, and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of MRI, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthineers, Shanghai, China
We had retrospectively enrolled 371 glioma patients in this study to develop an automated scheme to predict epilepsy in patients with WHO II-IV grade cerebral gliomas from multi-parametric MRI (mp-MRI). Gliomas tumor was segmented by a segmentation model trained with nnU-Net. Then a classification model based on ResNet-18 using segmented tumor region as anatomical attention was used to predict epilepsy from mp-MRI images. In the independent test cohort, the segmentation model achieved a mean dice of 0.899, while the classification model achieved an AUC of 0.890, better than the baseline ResNet-18 model with a test AUC of 0.783.
|
||
3189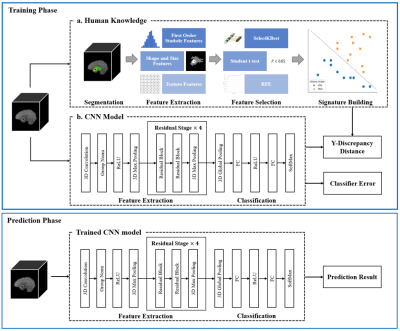 |
3 | Human Knowledge Guided Deep Learning with Multi-parametric MR Images for Glioma Grading
Yeqi Wang1,2, Longfei Li1,2, Cheng Li2, Hairong Zheng2, Yusong Lin1, and Shanshan Wang2
1School of Information Engineering, Zhengzhou University, Zhengzhou, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Automatic glioma grading based on magnetic resonance imaging (MRI) is crucial for appropriate clinical managements. Recently, Convolutional Neural Networks (CNNs)-based classification models have been extensively investigated. However, to achieve accurate glioma grading, tumor segmentation maps are typically required for these models to locate important regions. Delineating the tumor regions in 3D MR images is time-consuming and error-prone. Our target in this study is to develop a human knowledge guided CNN model for glioma grading without the reliance of tumor segmentation maps in clinical applications. Extensive experiments are conducted utilizing a public dataset and promising grading performance is achieved.
|
||
3190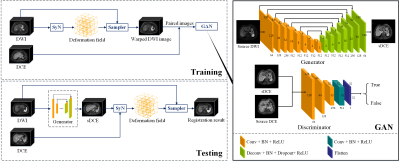 |
4 | Deep Learning-based Generative Adversarial Registration NETwork (GARNET) for Hepatocellular Carcinoma Segmentation: Multi-center Study
Hang Yu1, Rencheng Zheng1, Weibo Chen2, Ruokun Li3, Huazheng Shi4, Chengyan Wang5, and He Wang1
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Philips Healthcare, Shanghai, China, 3Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 4Shanghai Universal cloud imaging dignostic center, Shanghai, China, 5Human Phenome Institute, Fudan University, ShangHai, China
This study proposed a deep learning-based generative adversarial registration network (GARNET) for multi-contrast liver image registration and evaluated its value for hepatocellular carcinoma (HCC) segmentation. We used generative adversarial net (GAN) to synthesize images from diffusion-weighted imaging (DWI) to dynamic contrast-enhanced (DCE) and then applied for deformable registration on the synthesized DCE images. A total of 607 cirrhosis patients from 5 centers (401 HCC patients) were included in this study. We compared the proposed method with symmetric image normalization (SyN) registration and VoxelMorph. Experimental results demonstrated that GARNET improved the registration performances significantly and yielded better segmentation of HCC lesions.
|
||
3191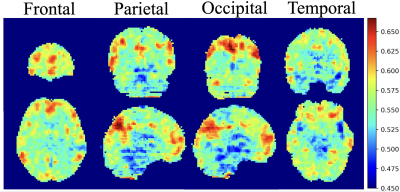 |
5 | Classification and visualization of chemo-brain in breast cancer survivors with deep residual and densely connected networks Video Not Available
Kai-Yi Lin1, Vincent Chin-Hung Chen2,3, Yuan-Hsiung Tsai2,4, and Jun-Cheng Weng1,3,5
1Department of Medical Imaging and Radiological Sciences, Graduate Institute of Artificial Intelligence, Chang Gung University, Taoyuan, Taiwan, 2School of Medicine, Chang Gung University, Taoyuan, Taiwan, 3Department of Psychiatry, Chang Gung Memorial Hospital, Chiayi, Taiwan, 4Department of Diagnostic Radiology, Chang Gung Memorial Hospital, Chiayi, Taiwan, 5Medical Imaging Research Center, Institute for Radiological Research, Chang Gung University and Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan
Our goal was to establish objective 3D deep learning models that differentiate cerebral alterations based on the effect of chemotherapy and to visualize the pattern that was recognized by our model. The average performance of SE-ResNet-50 models was accuracy of 80%, precision of 78%, and 70% recall, and the SE-DenseNet-121 model reached identical results with an average 80% accuracy, 86% precision, and 80% recall. The regions with the greatest contributions highlighted by the integrated gradients algorithm for differentiating chemo-brain were default mode and dorsal attention networks. We hope these results will be helpful in clinically tracking chemo-brain in the future.
|
||
3192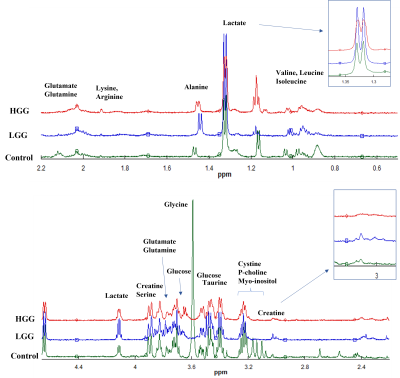 |
6 | HRMAS-NMR and Machine learning assisted untargeted Serum Metabolomics identified a panel of circulating biomarkers for detection of glioma. Video Permission Withheld
SAFIA FIRDOUS1,2, Zubair Nawaz3, Leo Ling Cheng4, and Saima Sadaf2
1Faculty of Rehabilitation and Allied Health Sciences, Riphah International University, Lahore, Pakistan, 2School of Biochemistry and Biotechnology, University of the Punjab, Lahore, Pakistan, 3Punjab University College of Information Technology, University of the Punjab, Lahore, Pakistan, 4Radiopahtological Unit, Massachusetts General Hospital, Boston, MA, United States
Metabolic alterations, crucial indicators of glioma development, can be used for detection of glioma before the appearance of fatal phenotype. We have compared the circulating metabolic fingerprints of glioma (n=26) and healthy controls (n=16) to identify a panel of biomarkers for detection of glioma. HRMAS-NMR spectra was obtained from two study groups and data was analysed by ML as well as chemometric methods (PCA and PLSDA). A panel of 38 metabolites was identified by three ML algorithms (logistics regression, extra tree classifier, & random forest), Wilcoxon test (p<0.05), and PLSDA (VIP score>1) which can serve as diagnostic biomarker of glioma.
|
||
3193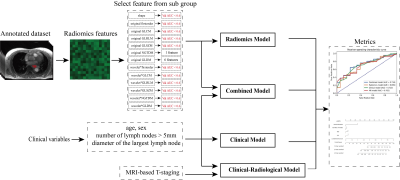 |
7 | Overall survival analysis of esophageal squamous cell carcinoma using MRI-based radiomics features
Yun Liu1, Chenglong Wang1, Funing Chu2, Jinrong Qu2, Xu Yan3, and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, The Affiliated Cancer Hospital of Zhengzhou University &Henan Cancer Hospital, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthineers, Shanghai, China
A total number of 439 patients with esophageal squamous cell carcinoma (ESCC) were enrolled in this study. All patients scanned using StarVIBE sequences. We split the data randomly into training and independent test cohort in a ratio of 7 to 3. We proposed a method for feature selection to find the most useful features for survival analysis. The radiomics score combined with clinical variables achieved the highest consistency in the prediction of disease-free survival (DFS) with a C-index value of 0.682 in the test cohort and overall survival (OS) with a C-index value of 0.691 in the test cohort.
|
||
3194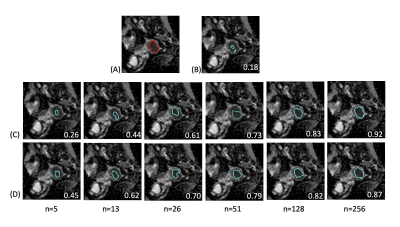 |
8 | Toward Universal Tumor Segmentation on Diffusion-Weighted MRI: Transfer Learning from Cervical Cancer to All Uterine Malignancies Video Permission Withheld
Yu-Chun Lin1, Yenpo Lin1, Yen-Ling Huang1, Chih-Yi Ho1, and Gigin Lin1,2
1Department of Medical Imaging and Intervention, Chang Gung Memorial Hospital at Linkou, Chang Gung Memorial Hospital, Taoyuan, Taiwan, 2Clinical Metabolomics Core Laboratory, Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan
This study retrospectively analyzed diffusion-weighted MRI in 320 patients with malignant uterine tumors (UT). A pretrained model was established for cervical cancer dataset. Transfer learning (TL) experiments were performed by adjusting fine-tuning layers and proportions of training data sizes. When using up to 50% of the training data, the TL models outperformed all the models. When the full dataset was used, the aggregated model exhibited the best performance, while the UT-only model exhibited the best in the UT dataset. TL of tumor segmentation on diffusion-weighted MRI for all uterine malignancy is feasible with limited case number.
|
||
3195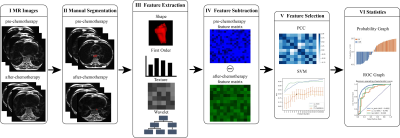 |
9 | Use delta-radiomics based on T2-TSE-BLADE MRI images to predict histopathological tumor regression grade in Locally Advanced Esophageal Cancer
Yun Liu1, Shuang Lu2, Chenglong Wang1, Xu Yan3, Jinrong Qu2, and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, The Affiliated Cancer Hospital of Zhengzhou University &Henan Cancer Hospital, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthineers, Shanghai, China
Histopathological tumor regression grade (TRG) has shown to be an important consideration for the choice of treatment plan in patients with esophagus cancer. Patients with same TNM stage may have different sensitivities to neoadjuvant chemotherapy (NAC). In this study, we proposed a new model to predict TRG grades by using the differences between radiomics features extracted from T2-TSE-BLADE images within 1 week before NAC and 3 to 4 weeks after NAC, but before surgery. This study enrolled 108 patients with esophageal cancer and underwent the mentioned MRI scans. In the test cohort, the proposed model achieved an AUC of 0.842.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.