Online Gather.town Pitches
Machine Learning & Artificial Intelligence IV
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
4329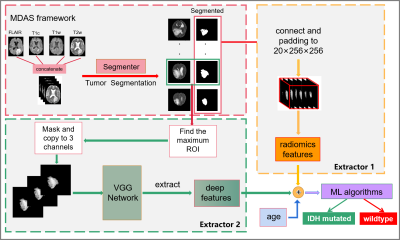 |
1 | Automated IDH genotype prediction pipeline using multimodal domain adaptive segmentation (MDAS) model
Hailong Zeng1, Lina Xu1, Zhen Xing2, Wanrong Huang2, Yan Su2, Zhong Chen1, Dairong Cao2, Zhigang Wu3, Shuhui Cai1, and Congbo Cai1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Imaging, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, ShenZhen, China
Mutation status of isocitrate dehydrogenase (IDH) in gliomas exhibits distinct prognosis. It poses challenges to jointly perform tumor segmentation and gene prediction directly using label-deprived multi-parametric MR images from clinics . We propose a novel multimodal domain adaptive segmentation (MDAS) framework, which derives unsupervised segmentation of tumor foci by learning data distribution between public dataset with labels and label-free targeted dataset. High-level features of radiomics and deep network are combined to manage IDH subtyping. Experiments demonstrate that our method adaptively aligns dataset from both domains with more tolerance toward distribution discrepancy during segmentation procedure and obtains competitive genotype prediction performance.
|
||
4330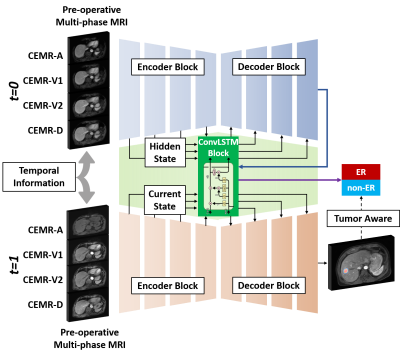 |
2 | Tumor Aware Temporal Deep Learning (TAP-DL) for Prediction of Early Recurrence in Hepatocellular Carcinoma Patients after Ablation using MRI
Yuze Li1, Chao An2, and Huijun Chen1
1Center for Biomedical Imaging Research, School of Medicine, Tsinghua University, Beijing, China, 2First Affiliated Hospital of Chinese PLA General Hospital, Beijing, China
Hepatocellular carcinoma patients after thermal ablation suffer high recurrence rate. In this study, we proposed a deep learning method to predict the early recurrence in these patients. Compared with other predictive models, two innovations were achieved in our study: 1) integrating interconnected tasks, i.e., tumor segmentation and tumor progression prediction, into a unified model to perform co-optimization; 2) using longitudinal images to take the therapy-induced changes into consideration to explore the temporal information. Results showed that our approach can simultaneously perform tumor segmentation and tumor progression prediction with higher performance than only doing any single one of them.
|
||
4331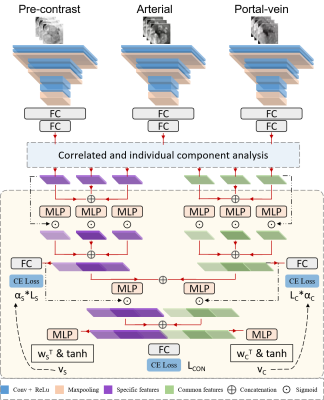 |
3 | Correlated and specific features fusion based on attention mechanism for grading hepatocellular carcinoma with Contrast-enhanced MR
Shangxuan Li1, Guangyi Wang2, Lijuan Zhang3, and Wu Zhou1
1School of Medical Information Engineering, Guangzhou University of Chinese Medicine, Guangzhou, China, 2Department of Radiology, Guangdong General Hospital, Guangzhou, China, 3Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Contrast-enhanced MR plays an important role in the characterization of hepatocellular carcinoma (HCC). In this work, we propose an attention-based common and specific features fusion network (ACSF-net) for grading HCC with Contrast-enhanced MR. Specifically, we introduce the correlated and individual components analysis to extract the common and specific features of Contrast-enhanced MR. Moreover, we propose an attention-based fusion module to adaptively fuse the common and specific features for better grading. Experimental results demonstrate that the proposed ACSF-net outperforms previously reported multimodality fusion methods for grading HCC. In addition, the weighting coefficient may have great potential for clinical interpretation.
|
||
4332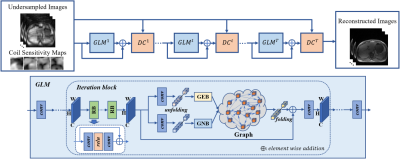 |
4 | Residual Non-local Attention Graph Learning (PNAGL) Neural Networks for Accelerating 4D-MRI
Bei Liu1, Huajun She1, Yufei Zhang1, Zhijun Wang1, and Yiping P. Du1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Residual non-local attention graph learning neural networks are proposed for accelerating 4D-MRI. Stack-of-star GRE radial sequence with self-navigator is used to acquire the data. We explore non-local self-similarity features in 4d-MR images by using residual non-local attention methods, and we use a graph convolutional network with an adaptive number of neighbor nodes to explore graph edge features. A global residual connection of graph learning model is used to further improve the performance. Through exploring non-local prior, the proposed method has the potential to be used in clinical applications such as MRI-guided real-time surgery.
|
||
4333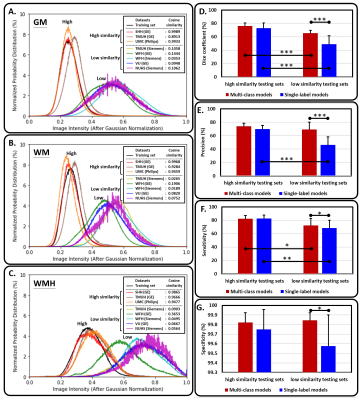 |
5 | A multi-output deep learning algorithm to improve brain lesion segmentation by enhancing the resistance of variabilities in tissue contrast
Yi-Tien Li1,2, Hsiao-Wen Chung3, and David Yen-Ting Chen4
1Neuroscience Research Center, Taipei Medical University, Taipei, Taiwan, 2Translational Imaging Research Center, Taipei Medical University Hospital, Taipei, Taiwan, 3Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan, 4Department of Medical Imaging, Taipei Medical University - Shuang Ho Hospital, New Taipei, Taiwan
We propose a multi-output segmentation approach, which incorporates other non-lesion brain tissue maps into the additional output layers to force the model to learn more about the lesion and tissue characteristics. We construct a cross-vendor study by training the white matter hyperintensities segmentation model on cases collected from one vendor and testing the model performance on eight different data sets. The model performance can be significantly improved, especially in testing sets which shows low image contrast similarity with training data, suggesting the feasibility of incorporating the non-lesion characteristics into segmentation model to enhance the resistance of cross-vendor image contrast variabilities.
|
||
4334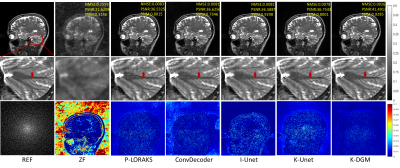 |
6 | k-Space Interpolation for Accelerated MRI Using Deep Generative Models
Zhuo-Xu Cui1, Sen Jia2, Zhilang Qiu2, Qingyong Zhu1, Yuanyuan Liu3, Jing Cheng2, Leslie Ying4, Yanjie Zhu2, and Dong Liang1
1Research Center for Medical AI, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3National Innovation Center for Advanced Medical Devices, Shenzhen, China, 4Department of Biomedical Engineering and the Department of Electrical Engineering, The State University of New York, Buffalo, NY, United States
k-space deep learning (DL) is emerging as an alternative to the conventional image domain DL for accelerated MRI. Typically, DL requires training on large amounts of data, which is unaccessible in clinical. This paper proposes to present an untrained k-space deep generative model (DGM) to interpolate missing data. Specifically, missing data is interpolated by a carefully designed untrained generator, of which the output layer conforms the MR image multichannel prior, while the architecture of other layers implicitly captures k-space statistics priors. Furthermore, we prove that the proposed method guarantees enough accuracy bounds for interpolated data under commonly used sampling patterns.
|
||
4335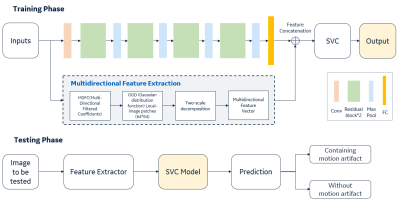 |
7 | A residual-spatial feature based MR motion artifact detection model with better generalization
Xiaolan Liu1, Yaan Ge1, Qingyu Dai1, and Kun Wang1
1GE Healthcare, Beijing, China Motion artifact in MRI images is the frequency existence in daily scanning and causes clinical distress. In this study, we proposed an automatic framework for MRI motion artifact detection using the residual features from neural network and spatial characteristics combination-based machine learning method, which can be applied to multiple body parts and sequences. High performance is achieved in validation with the accuracy of 97.6%. The comparison is performed with different representative methods and proving the effectiveness of the proposed architecture on limited dataset. |
||
4336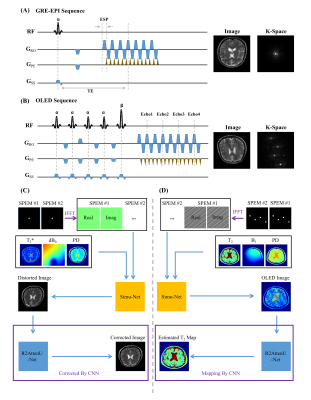 |
8 | High efficient Bloch simulation of MRI sequences based on deep learning
Haitao Huang1, Qinqin Yang1, Zhigang Wu2, Jianfeng Bao3, Jingliang Cheng3, Shuhui Cai1, and Congbo Cai1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2MSC Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China, 3Department of Magnetic Resonance Imaging, the First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, China
To overcome the difficulty of obtaining a large number of real training samples, the utilization of synthetic training samples based on Bloch simulation has become more and more popular in deep learning based MRI reconstruction. However, a large amount of Bloch simulation is usually very time-consuming even with the help of GPU. In this study, a simulation network that receives sequence parameters and contrast templates, was proposed to simulate MR images from different imaging sequences. The reliability and flexibility of the proposed method were verified by distortion correction for GRE-EPI images and T2 maps obtained with overlapping-echo detachment planar imaging.
|
||
4337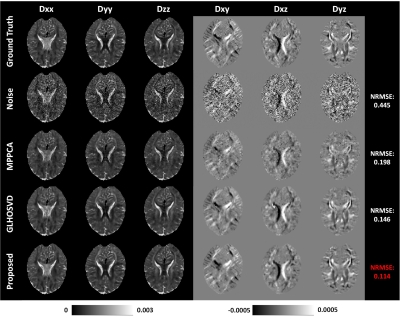 |
9 | Efficient six-direction DTI tensor estimation using model-based deep learning
Jialong Li1, Qiqi Lu1, Qiang Liu1, Yanqiu Feng1, and Xinyuan Zhang1
1School of Biomedical Engineering, Southern Medical University, Guangzhou, China
Diffusion tensor imaging (DTI) can noninvasively probe the tissue microstructure and characterize its anisotropic nature. The images carried with heavy diffusion-sensitizing gradients suffer from low SNR, and thus more than six diffusion-weighted images are required to improve the accuracy of parameter estimation against noise effect. We propose an efficient DTI model-based 3D-Unet (DTI-Unet) to predict high-quality diffusion tensor field and non-diffusion-weighted image from the noisy input. In our model, the input contains only six diffusion-weighted volumes and one b0 volume. Compared with the state-of-the-art denoising algorithms (MPPCA, GLHOSVD), our model performs better in image denoising and parameter estimation.
|
||
4338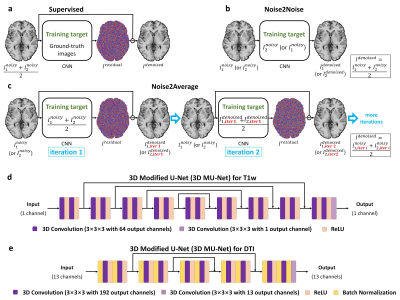 |
10 | Noise2Average: deep learning based denoising without high-SNR training data using iterative residual learning
Zihan Li1, Berkin Bilgic2,3, Ziyu Li4, Kui Ying5, Jonathan R. Polimeni2,3, Susie Huang2,3, and Qiyuan Tian2,3
1Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States, 3Harvard Medical School, Boston, MA, United States, 4Nuffield Department of Clinical Neurosciences, Oxford University, Oxford, United Kingdom, 5Department of Engineering Physics, Tsinghua University, Beijing, China
The requirement for high-SNR reference data reduces the feasibility of supervised deep learning-based denoising. Noise2Noise addresses this challenge by learning to map one noisy image to another repetition of the noisy image but suffers from image blurring resulting from imperfect image alignment and intensity mismatch for empirical MRI data. A novel approach, Noise2Average, is proposed to improve Noise2Noise, which employs supervised residual learning to preserve image sharpness and transfer learning for subject-specific training. Noise2Average is demonstrated effective in denoising empirical Wave-CAIPI MPRAGE T1-weighted data (R=3×3-fold accelerated) and DTI data and outperforms Noise2Noise and state-of-the-art BM4D and AONLM denoising methods.
|
||
4339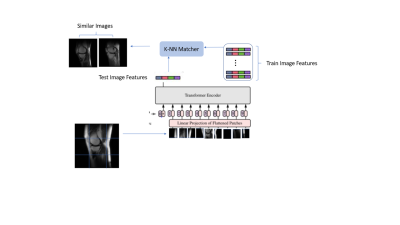 |
11 | Transformer based Self-supervised learning for content-based image retrieval Video Permission Withheld
Deepa Anand1, Chitresh Bhushan2, and Dattesh Dayanand Shanbhag1
1GE Healthcare, Bangalore, India, 2GE Global Research, Niskayuna, NY, United States
Diversity in training data encompassing variety of patient conditions is a recipe for the success of medical models based on DL. Ensuring diverse patient conditions is often impeded by the necessity to manually identify and include such cases, which is time-consuming and expensive. Here we propose a method of retrieving images similar to a handful of example images based on features learnt using self-supervised learning. We demonstrate the features learnt using SSL on transformer based networks are excellent feature learners which not only eliminates the need for annotation but enable accurate KNN based image retrieval matching the desired patient conditions.
|
||
4340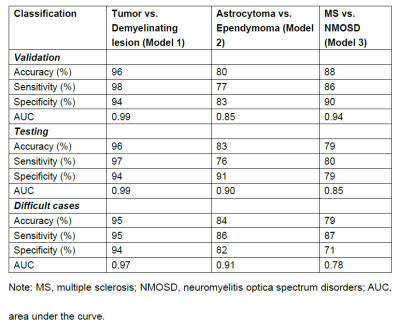 |
12 | Automated classification of intramedullary spinal cord tumors and inflammatory demyelinating lesions using deep learning Video Permission Withheld
Zhizheng Zhuo1, Jie Zhang1, Yunyun Duan1, Xianchang Zhang2, and Yaou Liu1
1Department of Radiology, Beijing Tiantan Hospital, Capital Medical University, Beijing, China, 2MR Collaboration, Siemens Healthineers Ltd, Beijing, China
A DL framework for the segmentation and classification of spinal cord lesions, including tumors (astrocytoma and ependymoma) and demyelinating diseases (MS and NMOSD), were developed and validated, with performance sometimes outperforming radiologists.
|
||
4341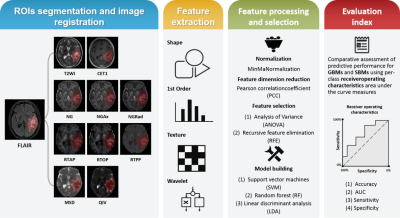 |
13 | Differentiation between glioblastoma and solitary brain metastasis using mean apparent propagator-MRI: a Radiomics analysis
Xiaoyue Ma1, Guohua Zhao1, Eryuan Gao1, Jinbo Qi1, Kai Zhao1, Ankang Gao1, Jie Bai1, Huiting Zhang2, Xu Yan2, Guang Yang3, and Jingliang Cheng1
1The Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2MR Scientific Marketing, Siemens Healthineers, Shanghai, China, 3Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China
Preoperative differentiation of glioblastomas and solitary brain metastases may contribute to more appropriate treatment plans and follow-up. However, routine MRI has a very limited ability to distinguish between the two. Mean apparent propagator (MAP)-MRI, as a representative of diffusion MRI technology, is effective in evaluating the complexity and inhomogeneity of the brain microstructure. We developed a series of radiomics models of MAP-MRI parametric maps, routine MRI, combined routine MRI, and combined MAP-MRI parametric maps to compare their performance in the identification of two tumors. Finally, a good performance with the combined MAP-MRI radiomics model was obtained.
|
||
4342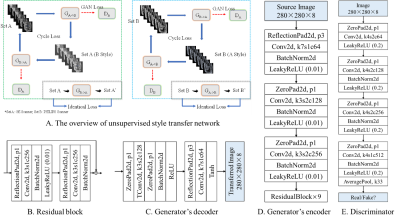 |
14 | Self-supervised learning for multi-center MRI harmonization without traveling phantoms: application for cervical cancer classification
Xiao Chang1, Xin Cai1, Yibo Dan2, Yang Song2, Qing Lu3, Guang Yang2, and Shengdong Nie1
1the Institute of Medical Imaging Engineering, University of Shanghai for Science and Technology, Shanghai, China, 2the Shanghai Key Laboratory of Magnetic Resonance, Department of Physics, East China Normal University, Shanghai, China, 3the Department of Radiology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
We proposed a self-supervised harmonization to achieve the generality and robustness of diagnostic models in multi-center MRI studies. By mapping the style of images from one center to another center, the harmonization without traveling phantoms was formalized as an unpaired image-to-image translation problem between two domains. The proposed method was demonstrated with pelvic MRI images from two different systems against two state-of-the-art deep-leaning (DL) based methods and one conventional method. The proposed method yields superior generality of diagnostic models by largely decreasing the difference in radiomics features and great image fidelity as quantified by mean structure similarity index measure (MSSIM).
|
||
4343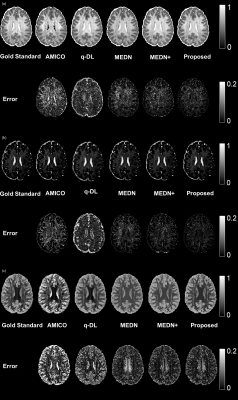 |
15 | A Microstructural Estimation Transformer with Sparse Coding for NODDI (METSCN)
Tianshu Zheng1, Yi-Cheng Hsu2, Yi Sun2, Yi Zhang1, Chuyang Ye3, and Dan Wu1
1Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, Zhejiang, China, Hangzhou, China, 2MR Collaboration, Siemens Healthineers Ltd., Shanghai, China, Shanghai, China, 3School of Information and Electronics, Beijing Institute of Technology, Beijing, China, Beijing, China Diffusion MRI (dMRI) models play an important role in characterizing tissue microstructures, commonly in the form of multi-compartmental biophysical models that are mathematically complex and highly non-linear. Fitting of these models with conventional optimization techniques is prone to estimation errors and requires dense sampling of q-space. Here we present a learning-based framework for estimating microstructural parameters in the NODDI model, termed Microstructure Estimation Transformer with Sparse Coding for NODDI (METSCN). We tested its performance with reduced q-space samples. Compared with the existing learning-based NODDI estimation algorithms, METSCN achieved the best accuracy, precision, and robustness. |
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.