Online Gather.town Pitches
Machine Learning & Artificial Intelligence I
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
3512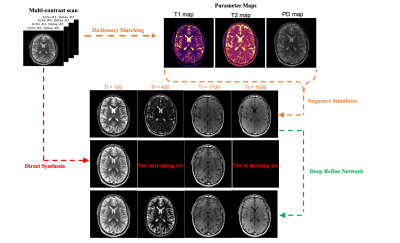 |
1 | Improving Synthetic MRI from Estimated Quantitative Maps with Deep Learning
Sidharth Kumar1 and Jonathan I Tamir1,2,3
1Electrical and Computer Engineering, University of Texas at Austin, Austin, TX, United States, 2Oden Institute for Computational Engineering and Sciences, University of Texas at Austin, Austin, TX, United States, 3Department of Diagnostic Medicine, University of Texas at Austin, Austin, TX, United States
Synthetic MRI has emerged as a tool for retrospectively generating contrast weightings from tissue parameter maps, but the generated contrasts can show mismatch due to unmodeled effects. We looked into the feasibility of refining arbitrary synthetic MRI contrasts using conditional GANs. To achieve this objective we trained a GAN on different experimentally obtained inversion recovery contrast images. As a proof of principle, the RefineNet is able to correct the contrast while losing out on finer structural details due to limited training data.
|
||
3513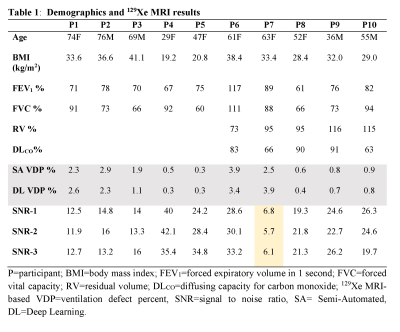 |
2 | Automated Quantification of Ventilation Defects and Heterogeneity in 3D Isotropic 129Xe MRI
Tuneesh K Ranota1, Fumin Guo2, Tingting Wu3, Matthew S Fox4, and Alexei Ouriadov3
1School of Biomedical Engineering, The University of Western Ontario, London, ON, Canada, 2Sunnybrook Research Institute, University of Toronto, Toronto, ON, Canada, 3Physics and Astronomy, The University of Western Ontario, London, ON, Canada, 4Physics and Astronomy, Lawson Health Research Institute, London, ON, Canada
Hyperpolarized 129Xe MRI provides a way to investigate and assess pulmonary diseases. To improve the performance of 129Xe MRI in lung imaging, a method for acquiring two VDP calculations using high-resolution 3D static ventilation imaging from FGRE combined with a key-hole method was proposed and demonstrated in participants with ventilation abnormalities. SNR was calculated as well as semi-automated VDP and fully automated VDP, which showed a strong positive linear correlation with a zero intercept and close to unity slop. This study confirms the feasibility of using isotropic-voxel 129Xe MRI acquired in a single breath-hold to study ventilation heterogeneity.
|
||
3514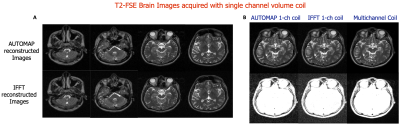 |
3 | Patch-based AUTOMAP image reconstruction of low SNR 1.5 T human brain MR k-space
Neha Koonjoo1,2, Bo Zhu1,2, Danyal Bhutto1,2,3, Suresh E Joel4, and Matthew S Rosen1,2,5
1Department of Radiology, A.A Martinos Center for Biomedical Imaging / MGH, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Department of Biomedical Engineering, Boston University, Boston, MA, United States, 4GE Healthcare, Bangalore, India, 5Department of Physics, Harvard University, Cambridge, MA, United States
AUTOMAP has proved itself to be robust to noise especially in the low SNR regimes, however due to the fully connected architecture of the input layer, its applicability to large matrix size datasets has been limited. Here we propose a patched-based trained network that enables the reconstruction of larger datasets. Low SNR single-channel volume coil brain images were acquired at 1.5T with different pulse sequences and reconstructed with the trained model. Results obtained show significant denoising potential. An increase in SNR of 1.5-fold as well as an increase in SSIM was also observed.
|
||
3515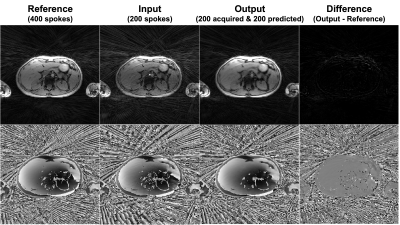 |
4 | A k-space transformer network for undersampled radial MRI
Chang Gao1,2, Shu-Fu Shih1,3, Vahid Ghodrati1,2, Paul Finn1,2, Peng Hu1,2, and Xiaodong Zhong4
1Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, United States, 2Department of Physics and Biology in Medicine, University of California Los Angeles, Los Angeles, CA, United States, 3Department of Bioengineering, University of California Los Angeles, Los Angeles, CA, United States, 4MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States
Deep learning-based undersampled MRI reconstruction generally requires the k-space data consistency term to constrain the output. However, this requires gridding onto a Cartesian basis for radial data, which slows down the training process profoundly and may even make it impractical. To avoid the repeated gridding process in training, we developed a transformer network to directly predict unacquired radial k-space spokes. The developed network was evaluated in vivo, accurately predicted the unacquired k-space spokes and generated better image intensity and less streaking artifacts compared to the undersampled images.
|
||
3516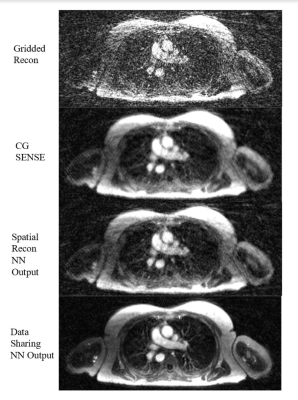 |
5 | Self-Supervised Deep Learning for Highly Accelerated 3D Ultrashort Echo Time Pulmonary MRI
Zachary Miller1 and Kevin Johnson1
1University of Wisconsin-Madison, Madison, WI, United States
The goal of this work is to reconstruct highly undersampled frames from dynamic Non-Cartesian acquisitions using deep learning. In this setting, supervised data is difficult to obtain due to organ motion necessitating self-supervision. The challenge is that acquisitions are often so data-starved that self-supervised reconstructions that use only spatial correlations fail to recover fine details. Here, we leverage correlations across time frames, and show that even when data is misaligned, it is possible to reconstruct highly accelerated frames using self-supervised methods. We demonstrate the feasibility of this technique by reconstructing end-inspiratory phase images from respiratory binned Pulmonary UTE acquisitions.
|
||
3517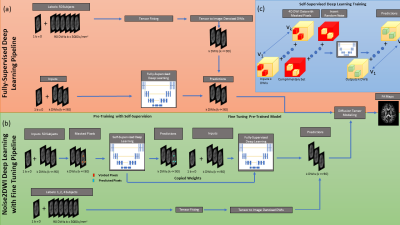 |
6 | Noise2DWI: Accelerating Diffusion Tensor Imaging with Self-Supervision and Fine Tuning
Phillip Martin1, Maria Altbach2,3, and Ali Bilgin1,2,3,4
1Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 3Medical Imaging, University of Arizona, Tucson, AZ, United States, 4Applied Mathematics, University of Arizona, Tucson, AZ, United States In this work we propose a novel algorithm of denoising accelerated diffusion weighted MRI (dMRI) acquisitions using deep learning and self-supervision. This method effectively enables the prediction of diffusion-weighted images (DWIs), without the need for large amounts of training data with high directional encodings. We demonstrate that accurate diffusion tensor metrics can be obtained with as few as 6 DWIs using only a few training datasets with high directional encodings. |
||
3518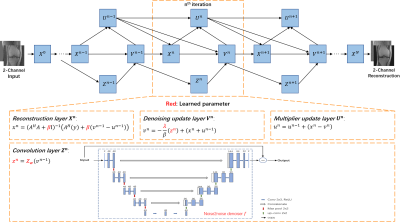 |
7 | Unsupervised Deep Unrolled Reconstruction Powered by Regularization by Denoising
Peizhou Huang1, Chaoyi Zhang2, Hongyu Li2, Ruiying Liu2, Xiaoliang Zhang1, Xiaojuan Li3, Dong Liang4, and Leslie Ying1,2
1Biomedical Engineering, State University of New York at Buffalo, Buffalo, NY, United States, 2Electrical Engineering, State University of New York at Buffalo, Buffalo, NY, United States, 3Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 4Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, CAS, Shenzhen, China
In this abstract, we propose a novel reconstruction method, named DURED-Net, that enables interpretable unsupervised learning for MR image reconstruction by combining an unsupervised denoising network and a plug-and-play method. Specifically, we incorporate the physics model into the denoising network using Regularization by Denoising (RED), and unroll the underlying optimization into a deep neural network. In addition, a sampling pattern was specially designed to facilitate unsupervised learning. Experiment results based on the knee fastMRI dataset exhibit marked improvements over the existing unsupervised reconstruction methods.
|
||
3519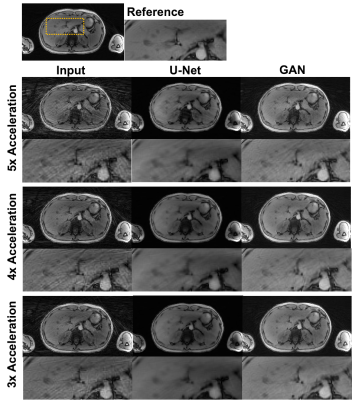 |
8 | Undersampling artifact reduction for free-breathing 3D stack-of-radial MRI based on a deep adversarial learning network
Chang Gao1,2, Vahid Ghodrati1,2, Shu-Fu Shih1,3, Holden H. Wu1,2,3, Yongkai Liu1,2, Marcel Dominik Nickel4, Thomas Vahle4, Brian Dale5, Victor Sai1, Ely Felker1, Qi Miao1, Xiaodong Zhong6, and Peng Hu1,2
1Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, United States, 2Department of Physics and Biology in Medicine, University of California Los Angeles, Los Angeles, CA, United States, 3Department of Bioengineering, University of California Los Angeles, Los Angeles, CA, United States, 4MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 5MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Cary, NC, United States, 6MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States
Undersampling is desired to reduce scan time but can cause streaking artifacts in stack-of-radial imaging. State-of-the-art deep neural networks such as the U-Net can be trained in a supervised manner to remove streaking artifacts but produce blurred images and loss of image details. Therefore, we developed and trained a 3D generative adversarial network to preserve perceptual image sharpness while removing streaking artifacts. The network used a combination of adversarial loss, L2 loss and structural similarity index loss. We demonstrated the feasibility of the proposed network for removing streaking artifacts and preserving perceptual image sharpness.
|
||
3520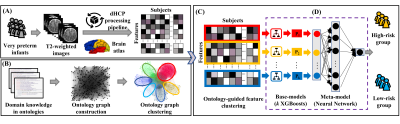 |
9 | A Ontology-guided Attribute Partitioning Ensemble Learning Model for Early Prediction of Cognitive Deficits using sMRI in Very Preterm Infants
Zhiyuan Li1,2, Hailong Li1,3, Nehal Parikh1,4, Lili He1,4, Adebayo Braimah1, and Jonathan Dillman1,5
1Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Electrical Engineering and Computer Science, University of Cincinnati, Cincinnati, OH, United States, 3The Perinatal Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 4Pediatrics, University of Cincinnati, Cincinnati, OH, United States, 5Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States
Between 35-40% of very preterm infants (≤32 weeks’ gestational age) develop cognitive deficits, thereby increasing their risk for poor educational, health, and social outcomes. Timely and accurate identification of infants at risk soon after birth is desirable for early intervention allocation. We proposed a novel Ontology-guided Attribute Partitioning Ensemble Learning (OAP-EL) model using quantitative structural MRI data obtained soon after birth to predict cognitive deficits at 2 years corrected age in very preterm infants.
|
||
3521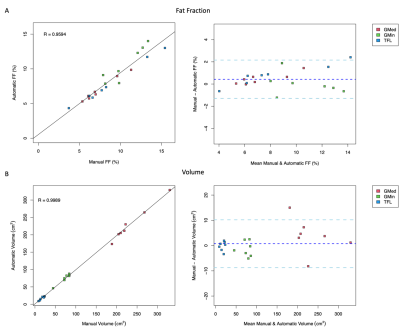 |
10 | AUTOMATIC EVALUATION OF HIP ABDUCTOR MUSCLE QUALITY IN HIP OSTEOARTHRITIS Video Not Available
Alyssa Bird1, Francesco Caliva1, Sharmila Majumdar1, Valentina Pedoia1, and Richard B Souza1
1University of California, San Francisco, San Francisco, CA, United States
The aim of this study was to reduce time-to-segmentation for analysis of muscle quality across a large volume of the hip abductors in individuals with hip osteoarthritis by developing an automatic segmentation network. The gluteus medius (GMED), gluteus minimus (GMIN), and tensor fasciae latae (TFL) were manually segmented on fat-water separated IDEAL MR images in 44 subjects. A 3D V-Net was trained, validated, and tested using these manually segmented image volumes and resulted in a mean Dice coefficient of 0.94, 0.87, and 0.91 for GMED, GMIN, and TFL. The automatic segmentation network demonstrated strong performance and provides drastically reduced time-to-segmentation.
|
||
3522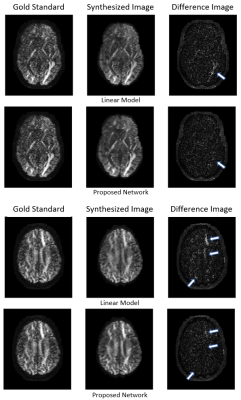 |
11 | Efficient Network for Diffusion-Weighted Image Interpolation and Accelerated Shell Sampling
Eric Y. Pierre1,2, Kieran O'Brien3, Thorsten Feiweier4, Josef Pfeuffer4, and Daniel Staeb2
1The Florey Institute of Neuroscience, Melbourne, Australia, 2MR Research Collaborations, Siemens Healthcare Pty Ltd, Bayswater, Australia, 3MR Research Collaborations, Siemens Healthcare Pty Ltd, Brisbane, Australia, 4MR Applications Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany We propose an efficient, densely connected network to synthesize unacquired DW-volumes from other acquired DW-directions and b=0 mm2/s volumes, allowing acceleration of high-angular DWI shell acquisition by skipping some DW-directions altogether. For training, we used a high-quality dataset of 20 HCP subjects with 90 DW-directions per subject at both b=1000 mm2/s and 3000 mm2/s. 40 HCP subjects were used for validation. 30 DW-directions were selected for input to reconstruct the other 60 missing target DW-directions. Comparison with a linear-interpolation benchmark show improved fidelity of synthesized DW-volumes and FOD maps to a gold standard acquisition, for both b-values.
|
||
3523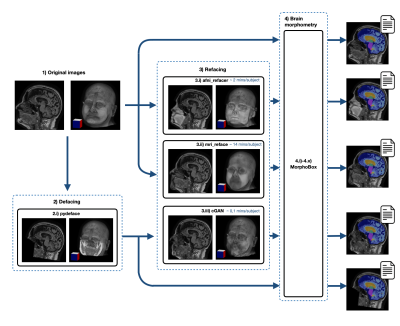 |
12 | Preserving Privacy While Maintaining Consistent Postprocessing Results: Fast and Effective Anonymous Refacing using a 3D cGAN
Nataliia Molchanova1,2, Bénédicte Maréchal1,2,3, Jean-Philippe Thiran2,3, Tobias Kober1,2,3, Jonas Richiardi3, and Till Huelnhagen1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Signal Processing Laboratory (LTS 5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland
We propose a novel refacing technique employing a 3D conditional generative adversarial network to allow protecting subject privacy while maintaining consistent post-processing results. We evaluate the method compared to current refacing techniques using brain morphometry as an example. Results show that the proposed method compromises brain morphometry results to a lesser extent than existing methods while showing lower similarity of the final image to the original one, hence suggesting an improved privacy protection. We conclude that the proposed method represents a fast and viable alternative for image data de-identification compared to currently existing methods.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.