Online Gather.town Pitches
Machine Learning/Artificial Intelligence III
Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting • 07-12 May 2022 • London, UK

| Booth # | ||||
|---|---|---|---|---|
3914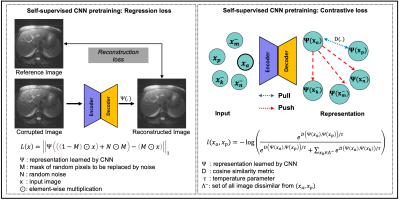 |
1 | Learning to segment with limited annotations: Self-supervised pretraining with Regression and Contrastive loss in MRI
Lavanya Umapathy1,2, Zhiyang Fu1,2, Rohit Philip2, Diego Martin3, Maria Altbach2,4, and Ali Bilgin1,2,4,5
1Department of Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Department of Medical Imaging, University of Arizona, Tucson, AZ, United States, 3Department of Radiology, Houston Methodist Hospital, Houston, TX, United States, 4Department of Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 5Program in Applied Mathematics, University of Arizona, Tucson, AZ, United States
The availability of large unlabeled datasets compared to labeled ones motivate the use of self-supervised pretraining to initialize deep learning models for subsequent segmentation tasks. We consider two pre-training approaches for driving a CNN to learn different representations using: a) a reconstruction loss that exploits spatial dependencies and b) a contrastive loss that exploits semantic similarity. The techniques are evaluated in two MR segmentation applications: a) liver and b) prostate segmentation in T2-weighted images. We observed that CNNs pretrained using self-supervision can be finetuned for comparable performance with fewer labeled datasets.
|
||
3915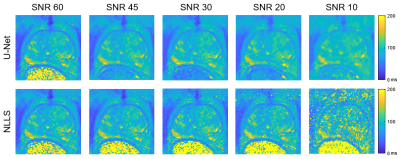 |
2 | T2 Mapping of the Prostate with a Convolutional Neural Network
Sara L Saunders1,2, Mitchell J Gross2, Gregory J Metzger1, and Patrick J Bolan1
1Center for MR Research / Radiology, University of Minnesota, MINNEAPOLIS, MN, United States, 2Biomedical Engineering, University of Minnesota, Minneapolis, MN, United States
This work compares T2 maps calculated from multi-echo spin-echo MR images using a conventional non-linear least squares (NLLS) fitting method to those constructed with a U-Net, a type of convolutional neural network. The performance of the U-Net and NLLS methods was compared in two retrospectively simulated experiments with a) reduced echo train lengths and b) decreased SNR to emulate accelerated acquisitions. The U-Net generally gave higher accuracy than NLLS fitting, with the trade-off of a modest increase of blurring of the resultant T2 maps.
|
||
3916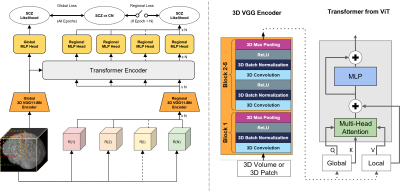 |
3 | Classification of Schizophrenia with Structural MRI Using 3D-MIGR Transformer
Ye Tian1, Junhao Zhang1, Xuemin Zhu1, Pin-Yu Lee1, Vishwanatha Mitnala Rao1, Zhuoyao Xin1, Andrew F Laine2, Scott A Small3, and Jia Guo4
1Columbia University, New York, NY, United States, 2Biomedical Engineering, Columbia University, New York, NY, United States, 3Neurology, Columbia University, New York, NY, United States, 4Psychiatry, Columbia University, New York, NY, United States To detect distinctive structural abnormalities of schizophrenia early in magnetic resonance imaging (MRI) data, we propose a 3D Medical Image Global-Regional (3D-MIGR) Transformer with a VGG11BN backbone followed by a global-regional transformer encoder, which outperforms state-of-the-art models. We trained and tested our model on 887 pre-processed structural whole-head (WH) T1W 3D images from 3 datasets with similar acquisition parameters. 3D-MIGR Transformer improves AUROC to 0.990 and demonstrates strong generality. We found that combining volume-level contextual information with patch-level features enhances performance and allows us to identify ventricle areas as the most informative regions in regional schizophrenia likelihood visualization. |
||
3917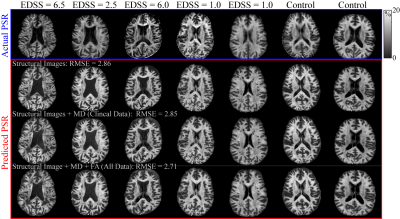 |
4 | Predicting Selective Inversion Recovery Myelin Images from Routine Clinical Images and a Simple Linear Model
Nicholas John Sisco1, Francesca Bagnato2, Ashley M Stokes1, and Richard D Dortch1
1Division of Neuroimaging Researc, Barrow Neurological Institute, Phoenix, AZ, United States, 2Neurology Department, Vanderbilt University Medical Center, Nashville, TN, United States Using a simple linear model, we predicted myelin-specific PSR maps from clinically available images. We trained this linear model on various combinations of routine images, including gray-matter standardized T1- and T2-weighted images and diffusion maps. The model reasonably predicted PSR values from structural images alone, although model performance was improved by the addition of diffusion parameters. In the future, this model may enable researchers and clinicians to assess myelin status without the need for specialized myelin imaging sequences. In addition, myelin maps could be obtained retrospectively in large imaging databases without myelin specific methods. |
||
3918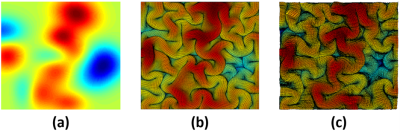 |
5 | Brain Growth and Folding Processes Using Deep Neural Networks
Yanchen Guo1, Poorya Chavoshnejad2, Mir Jalil Razavi2, and Weiying Dai1
1Computer Science, State University of New York at Binghamton, Binghamton, NY, United States, 2Mechanical Engineering, State University of New York at Binghamton, Binghamton, NY, United States Finite Element (FE)-based mechanical models can simulate the brain growth and folding process. But they are time consuming due to the large number of nodes in a real human brain and the reverse process to the initial smooth brain surfaces is difficult because it is not invertible problem. Here, we demonstrate a proof-of-concept that deep-learning neural networks (DNN) can learn the growth and folding process of human brain in forward and reverse directions and can predict/retrieve the developed/primary folding patterns in a very fast speed. |
||
3919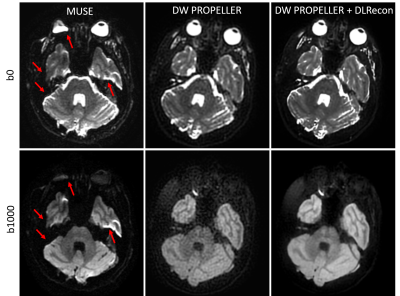 |
6 | Robust Diffusion-Weighted Imaging with Deep Learning-Based DW PROPELLER Reconstruction
Xinzeng Wang1, Ali Ersoz2, Daniel Litwiller3, Jingfei Ma4, Jason Stafford4, and Ersin Bayram1
1GE Healthcare, Houston, TX, United States, 2GE Healthcare, Waukesha, WI, United States, 3GE Healthcare, Denver, CO, United States, 4MD Anderson Cancer Center, Houston, TX, United States
Compared to DW-EPI, DW PROPELLER is less sensitive to susceptibility, chemical shift, and motion, and thus shows better image quality in areas, such as skull base, head-neck and pelvis. However, the SNR and in-plane resolution of DW PROPELLER images are often inferior to DW-EPI and often requires a longer scan time to compensate for this. In this work, we evaluated a deep-learning reconstruction method to improve the SNR of in-plane resolution of DW PROPELLER images without increasing acquisition time.
|
||
3920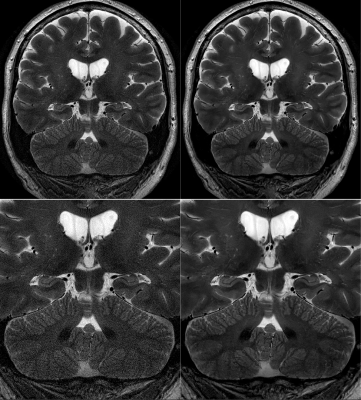 |
7 | Improving Motion-Robust Structural Imaging at 7T with Deep Learning-Based PROPELLER Reconstruction
Daniel V Litwiller1, Xinzeng Wang2, R Marc Lebel3, Baolian Yang4, Jeffrey McGovern4, Brian Burns5, and Suchandrima Banerjee6
1GE Healthcare, Denver, CO, United States, 2GE Healthcare, Houston, TX, United States, 3GE Healthcare, Calgary, AB, Canada, 4GE Healthcare, Waukesha, WI, United States, 5GE Healthcare, Olympia, WA, United States, 6GE Healthcare, Menlo Park, CA, United States
The high sensitivity of MRI at 7T enables brain imaging with unprecedented spatial resolution, which can be important to the assessment of a variety of neurological disorders, such as multiple sclerosis, epilepsy, and neurodegenerative disease. With sub-millimeter voxel dimensions, and prolonged acquisition times, however, sensitivity to motion and pulsatility is increased dramatically. This increased sensitivity to motion can be managed with techniques like PROPELLER. Here, we present an initial assessment of a deep learning-based image reconstruction for high-resolution, 7T PROPELLER, and evaluate its ability to improve signal-to-noise ratio, and anatomical conspicuity, without increasing scan time.
|
||
3921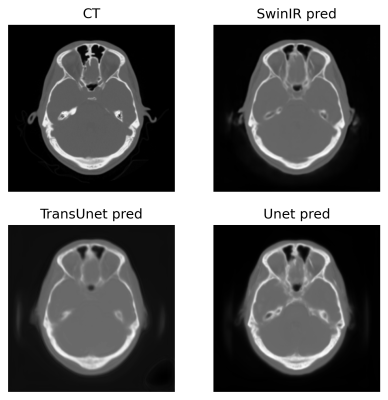 |
8 | Exploration of vision transformer models in medical images synthesis
Weijie Chen1, Seyed Iman Zare Estakhraji2, and Alan B McMillan3
1Department of Electrical and Computer Engineering, University of Wisconsin-Madison, Madison, WI, United States, 2Department of Radiology, University of Wisconsin-Madison, Madison, WI, United States, 3Department of Biomedical Engineering, Department of Radiology and Medical Physics, Madison, WI, United States Applications such as PET/MR and MR-only Radiotherapy Planning need the capability to derive a CT-like image from MRI inputs to enable accurate attenuation correction and dose estimation. More recently, transformer models have been proposed for computer vision applications. Models in the transformer family discard traditional convolution-based network structures and emphasize the importance of non-local information yielding potentially more realistic outputs. To evaluate the performance of SwinIR, TransUet, and Unet. After comparing results visually and quantitatively, the SwinIR models and TransUnet models appear to provide higher-quality synthetic CT scans compared to the conventional Unet. |
||
3922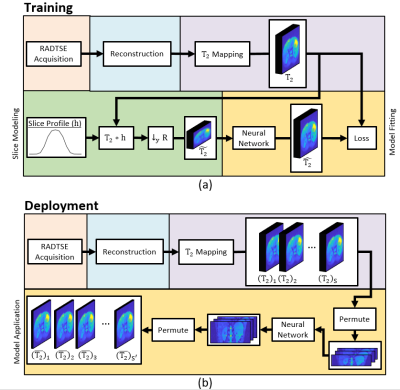 |
9 | Deep learning-based slice resolution for improved slice coverage in abdominal T2 mapping
Eze Ahanonu1, Zhiyang Fu1,2, Kevin Johnson2, Rohit Philip1, Diego R Martin3, Maria Altbach2,4, and Ali Bilgin1,2,4
1Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Medical Imaging, University of Arizona, Tucson, AZ, United States, 3Radiology, Houston Methodist Hospital, Houston, TX, United States, 4Biomedical Engineering, University of Arizona, Tucson, AZ, United States This study presents a deep learning based technique for slice super resolution in RADTSE T2 mapping. The proposed method may be used to accelerate full liver imaging, while maintaining sufficient through-plane resolution for detection of small pathologies. |
||
3923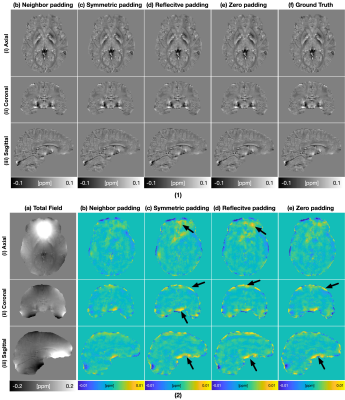 |
10 | Improved Padding in CNNs for Quantitative Susceptibility Mapping
Juan Liu1
1Yale University, NEW HAVEN, CT, United States
Recently, deep learning approaches have been proposed for QSM processing - background field removal, field-to-source inversion, and single-step QSM reconstruction. In these tasks, the networks usually take local fields or total fields as inputs, which have valid voxels within volume of interests (VOIs) and invalid voxels outside of VOIs. CNNs fail to consider this spatial information when using spatial invariant filters and conventional padding mechanism, which could introduce spatial artifacts in the QSM results. Here, we propose an improved padding technique utilizing neighboring valid voxels of invalid voxels to estimate the invalid voxels in feature maps of CNNs.
|
||
3924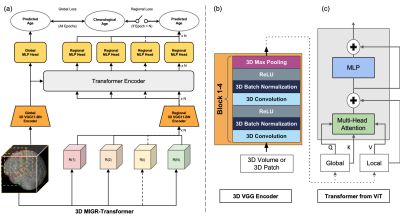 |
11 | Increased Brain Age Gap Estimate (BrainAGE) in APOE4 carriers with Alzheimer’s disease
Xuemin Zhu1, Pin-Yu Lee2, Ye Tian2, Andrew F. Laine3, Tal Nuriel4, and Jia Guo5
1Zuckerman Institute, New York, NY, United States, 2Columbia University, New York, NY, United States, 3Biomedical Engineering, Columbia University, New York, NY, United States, 4Taub Institute, New York, NY, United States, 5Psychiatry, Columbia University, New York, NY, United States
Recent evidence suggests APOE4 is the most potent genetic risk factor for late-onset Alzheimer’s disease. However, researchers have not treated its impact on brain aging. We proposed a 3D Medical Image Global Regional (MIGR) Transformer to determine whether APOE4 affects brain age gap estimate using MRI. We achieved a mean absolute error of 4.71 on brain age estimate in a large healthy dataset (n = 2852) with an age range of 18-97 years from 13 public datasets. We investigated the brainAGE on three stages of Alzheimer’s disease. We found an accelerating trend of brain aging for APOE4 carriers.
|
||
3925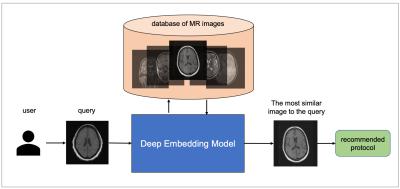 |
12 | MRI protocol recommendation using deep metric learning
Mohamad Abdi1, Yu Zhao1, Sepehr Farhand1, Ke Zeng1, Mahesh Ranganath1, Yoshihisa Shinagawa1, and Gerardo Valadez Hermosillo1
1Siemens Healthineers, Malvern, PA, United States
MRI requires careful design of imaging protocols and parameters to optimally assess a particular region of the body and/or pathological process. Selection of acquisition parameters is a challenging task because (a) the relationship between the acquisition parameters and the image features is typically non-trivial, and (b) not all users have the leverage to optimize their imaging protocols. To help users overcome these challenges and elevate the user experience, a deep metric learning tool was developed as a recommendation system for automatic candidate generation of imaging protocols. The feasibility of the model is evaluated using 3-dimensional brain MR images.
|
||
3926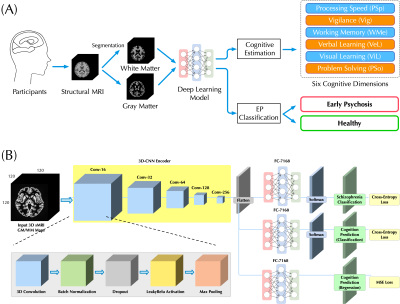 |
13 | Bridging Structural MRI with Cognitive Function for Individual Level Classification of Early Psychosis via Deep Learning
Yang Wen1,2,3, Chuan Zhou2, Leiting Chen2, Yu Deng4, Martine Cleusix5, Raoul Jenni5, Philippe Conus6, Kim Q. Do5, and Lijing Xin1,3
1Animal imaging and technology core (AIT), Center for Biomedical Imaging (CIBM), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 2University of Electronic Science and Technology of China, Chengdu, China, 3Laboratory of Functional and Metabolic Imaging, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4Department of Biomedical Engineering, King's College London, London, United Kingdom, 5Center for Psychiatric Neuroscience, Department of Psychiatry, Centre Hospitalier Universitaire Vaudois (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland, 6Service of General Psychiatry, Department of Psychiatry, Centre Hospitalier Universitaire Vaudois (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland
The aims of this study were: (1) to test the feasibility of using a deep learning model with 7T sMRI as an input to predict cognition levels (CLs) at the single-subject level, and (2) to investigate whether the inclusion of CLs estimation could facilitate the classification for early psychosis (EP) patients and healthy controls (HCs). Promising accuracy was achieved in estimating CLs and the inclusion provides considerable classification improvement. Fivefold cross-validating experiments demonstrated higher classification AUC-ROC scores over published methods. Therefore, deep learning can be used to estimate CLs and CL estimation improves the classification performance of EP.
|
||
3927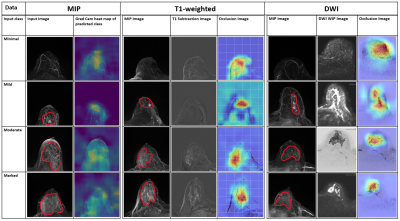 |
14 | Detection and prediction of background parenchymal enhancement on breast MRI using deep learning
Badhan Kumar Das1,2, Lorenz A. Kapsner1, Sabine Ohlmeyer1, Frederik B. Laun1, Andreas Maier2, Michael Uder1, Evelyn Wenkel1, Sebastian Bickelhaupt1, and Andrzej Liebert1
1Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nürnberg (FAU), Erlangen, Germany, 2Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander University Erlangen-Nürnberg (FAU), Erlangen, Germany The purpose of this work was to automatically classify BPE using T1-weighted subtraction volumes and diffusion-weighted imaging volumes in breast MRI. The dataset consisted of 621 routine breast MRI examination acquired at University Hospital Erlangen. 2D MIP and 3D T1-subtraction volumes were used for the automatic detection of BPE classes. Multi-b-value DWI (up to1500s/mm2) DWI images were used for automatic prediction. ResNet and DenseNet models were used for 2D and 3D data respectively. The study demonstrated an AUROC of 0.8107 on the test set using the T1-subtraction volumes. With DWI volumes, a slightly decreased AuROC of 0.78 was achieved. |
||
3928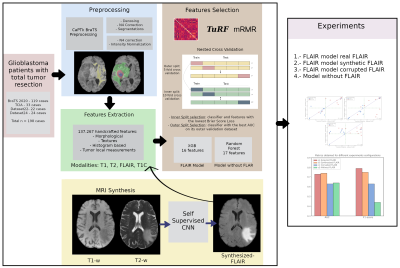 |
15 | Synthetic MRI aids in glioblastoma survival prediction
Rafael Navarro-González1, Elisa Moya-Sáez1, Rodrigo de Luis-García1, Santiago Aja-Fernández1, and Carlos Alberola-López1
1University of Valladolid, Valladolid, Spain
Radiomics systems for survival prediction in glioblastoma multiforme could enhance patient management, personalizing its treatment and obtaining better outcomes. However, these systems are data-demanding multimodality images. Thus, synthetic MRI could improve radiomics systems by retrospectively completing databases or replacing artifacted images. In this work we analyze the replacement of an acquired modality by a synthesized counterversion for predicting survival with an independent radiomic system. Results prove that a model fed with the synthesized modality achieves similar performance compared to using the acquired modality, and better performance than using a corrupted modality or a model trained from scratch without this modality.
|
||
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.